et al.
et al.
et al.
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
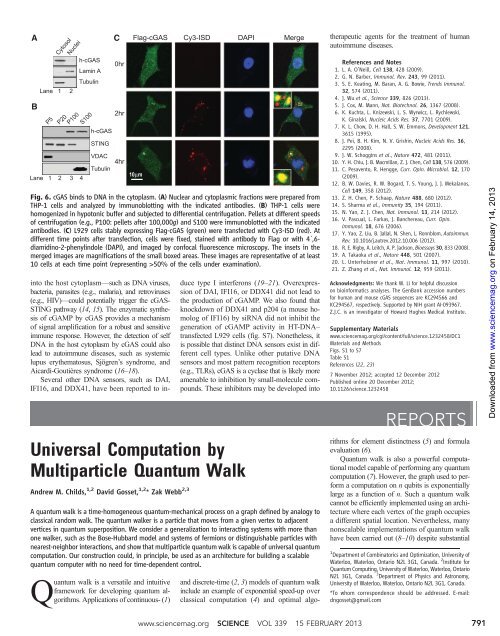
Fig. 6. cGAS binds to DNA in the cytoplasm. (A) Nuclear and cytoplasmic fractions were prepared from<br />
THP-1 cells and an<strong>al</strong>yzed by immunoblotting with the indicated antibodies. (B) THP-1 cells were<br />
homogenized in hypotonic buffer and subjected to differenti<strong>al</strong> centrifugation. Pell<strong>et</strong>s at different speeds<br />
of centrifugation (e.g., P100: pell<strong>et</strong>s after 100,000g) and S100 were immunoblotted with the indicated<br />
antibodies. (C) L929 cells stably expressing Flag-cGAS (green) were transfected with Cy3-ISD (red). At<br />
different time points after transfection, cells were fixed, stained with antibody to Flag or with 4´,6diamidino-2-phenylindole<br />
(DAPI), and imaged by confoc<strong>al</strong> fluorescence microscopy. The ins<strong>et</strong>s in the<br />
merged images are magnifications of the sm<strong>al</strong>l boxed areas. These images are representative of at least<br />
10 cells at each time point (representing >50% of the cells under examination).<br />
into the host cytoplasm—such as DNA viruses,<br />
bacteria, parasites (e.g., m<strong>al</strong>aria), and r<strong>et</strong>roviruses<br />
(e.g., HIV)—could potenti<strong>al</strong>ly trigger the cGAS-<br />
STING pathway (14, 15). The enzymatic synthesis<br />
of cGAMP by cGAS provides a mechanism<br />
of sign<strong>al</strong> amplification for a robust and sensitive<br />
immune response. However, the d<strong>et</strong>ection of self<br />
DNA in the host cytoplasm by cGAS could <strong>al</strong>so<br />
lead to autoimmune diseases, such as systemic<br />
lupus erythematosus, Sjögren’s syndrome, and<br />
Aicardi-Goutières syndrome (16–18).<br />
Sever<strong>al</strong> other DNA sensors, such as DAI,<br />
IFI16, and DDX41, have been reported to in-<br />
Univers<strong>al</strong> Computation by<br />
Multiparticle Quantum W<strong>al</strong>k<br />
Andrew M. Childs, 1,2 David Goss<strong>et</strong>, 1,2 * Zak Webb 2,3<br />
duce type I interferons (19–21). Overexpression<br />
of DAI, IFI16, or DDX41 did not lead to<br />
the production of cGAMP. We <strong>al</strong>so found that<br />
knockdown of DDX41 and p204 (a mouse homolog<br />
of IFI16) by siRNA did not inhibit the<br />
generation of cGAMP activity in HT-DNA–<br />
transfected L929 cells (fig. S7). Non<strong>et</strong>heless, it<br />
is possible that distinct DNA sensors exist in different<br />
cell types. Unlike other putative DNA<br />
sensors and most pattern recognition receptors<br />
(e.g., TLRs), cGAS is a cyclase that is likely more<br />
amenable to inhibition by sm<strong>al</strong>l-molecule compounds.<br />
These inhibitors may be developed into<br />
A quantum w<strong>al</strong>k is a time-homogeneous quantum-mechanic<strong>al</strong> process on a graph defined by an<strong>al</strong>ogy to<br />
classic<strong>al</strong> random w<strong>al</strong>k. The quantum w<strong>al</strong>ker is a particle that moves from a given vertex to adjacent<br />
vertices in quantum superposition. We consider a gener<strong>al</strong>ization to interacting systems with more than<br />
onew<strong>al</strong>ker,suchastheBose-Hubbardmodelandsystems of fermions or distinguishable particles with<br />
nearest-neighbor interactions, and show that multiparticle quantum w<strong>al</strong>k is capable of univers<strong>al</strong> quantum<br />
computation. Our construction could, in principle, be used as an architecture for building a sc<strong>al</strong>able<br />
quantum computer with no need for time-dependent control.<br />
Quantum w<strong>al</strong>k is a versatile and intuitive<br />
framework for developing quantum <strong>al</strong>gorithms.<br />
Applications of continuous- (1)<br />
and discr<strong>et</strong>e-time (2, 3) models of quantum w<strong>al</strong>k<br />
include an example of exponenti<strong>al</strong> speed-up over<br />
classic<strong>al</strong> computation (4) and optim<strong>al</strong> <strong>al</strong>go-<br />
therapeutic agents for the treatment of human<br />
autoimmune diseases.<br />
References and Notes<br />
1. L. A. O’Neill, Cell 138, 428 (2009).<br />
2. G. N. Barber, Immunol. Rev. 243, 99 (2011).<br />
3. S. E. Keating, M. Baran, A. G. Bowie, Trends Immunol.<br />
32, 574 (2011).<br />
4. J. Wu <strong>et</strong> <strong>al</strong>., Science 339, 826 (2013).<br />
5. J. Cox, M. Mann, Nat. Biotechnol. 26, 1367 (2008).<br />
6. K. Kuchta, L. Knizewski, L. S. Wyrwicz, L. Rychlewski,<br />
K. Gin<strong>al</strong>ski, Nucleic Acids Res. 37, 7701 (2009).<br />
7. K. L. Chow, D. H. H<strong>al</strong>l, S. W. Emmons, Development 121,<br />
3615 (1995).<br />
8. J. Pei, B. H. Kim, N. V. Grishin, Nucleic Acids Res. 36,<br />
2295 (2008).<br />
9. J. W. Schoggins <strong>et</strong> <strong>al</strong>., Nature 472, 481 (2011).<br />
10. Y. H. Chiu, J. B. Macmillan, Z. J. Chen, Cell 138, 576 (2009).<br />
11. C. Pesavento, R. Hengge, Curr. Opin. Microbiol. 12, 170<br />
(2009).<br />
12. B. W. Davies, R. W. Bogard, T. S. Young, J. J. Mek<strong>al</strong>anos,<br />
Cell 149, 358 (2012).<br />
13. Z. H. Chen, P. Schaap, Nature 488, 680 (2012).<br />
14. S. Sharma <strong>et</strong> <strong>al</strong>., Immunity 35, 194 (2011).<br />
15. N. Yan, Z. J. Chen, Nat. Immunol. 13, 214 (2012).<br />
16. V. Pascu<strong>al</strong>, L. Farkas, J. Banchereau, Curr. Opin.<br />
Immunol. 18, 676 (2006).<br />
17. Y. Yao, Z. Liu, B. J<strong>al</strong>l<strong>al</strong>, N. Shen, L. Ronnblom, Autoimmun.<br />
Rev. 10.1016/j.autrev.2012.10.006 (2012).<br />
18. R. E. Rigby, A. Leitch, A. P. Jackson, Bioessays 30, 833 (2008).<br />
19. A. Takaoka <strong>et</strong> <strong>al</strong>., Nature 448, 501 (2007).<br />
20. L. Unterholzner <strong>et</strong> <strong>al</strong>., Nat. Immunol. 11, 997 (2010).<br />
21. Z. Zhang <strong>et</strong> <strong>al</strong>., Nat. Immunol. 12, 959 (2011).<br />
Acknowledgments: W<strong>et</strong>hankW.Liforhelpfuldiscussion<br />
on bioinformatics an<strong>al</strong>yses. The GenBank accession numbers<br />
for human and mouse cGAS sequences are KC294566 and<br />
KC294567, respectively. Supported by NIH grant AI-093967.<br />
Z.J.C. is an investigator of Howard Hughes Medic<strong>al</strong> Institute.<br />
Supplementary Materi<strong>al</strong>s<br />
www.sciencemag.org/cgi/content/full/science.1232458/DC1<br />
Materi<strong>al</strong>s and M<strong>et</strong>hods<br />
Figs. S1 to S7<br />
Table S1<br />
References (22, 23)<br />
7 November 2012; accepted 12 December 2012<br />
Published online 20 December 2012;<br />
10.1126/science.1232458<br />
REPORTS<br />
rithms for element distinctness (5) and formula<br />
ev<strong>al</strong>uation (6).<br />
Quantum w<strong>al</strong>k is <strong>al</strong>so a powerful computation<strong>al</strong><br />
model capable of performing any quantum<br />
computation (7). However, the graph used to perform<br />
a computation on n qubits is exponenti<strong>al</strong>ly<br />
large as a function of n. Such a quantum w<strong>al</strong>k<br />
cannot be efficiently implemented using an architecture<br />
where each vertex of the graph occupies<br />
a different spati<strong>al</strong> location. Nevertheless, many<br />
nonsc<strong>al</strong>able implementations of quantum w<strong>al</strong>k<br />
have been carried out (8–10) despite substanti<strong>al</strong><br />
1<br />
Department of Combinatorics and Optimization, University of<br />
Waterloo, Waterloo, Ontario N2L 3G1, Canada. 2 Institute for<br />
Quantum Computing, University of Waterloo, Waterloo, Ontario<br />
N2L 3G1, Canada. 3 Department of Physics and Astronomy,<br />
University of Waterloo, Waterloo, Ontario N2L 3G1, Canada.<br />
*To whom correspondence should be addressed. E-mail:<br />
dngoss<strong>et</strong>@gmail.com<br />
www.sciencemag.org SCIENCE VOL 339 15 FEBRUARY 2013 791<br />
on February 14, 2013<br />
www.sciencemag.org<br />
Downloaded from