Transcriptional Characterization of Glioma Neural Stem Cells Diva ...
Transcriptional Characterization of Glioma Neural Stem Cells Diva ...
Transcriptional Characterization of Glioma Neural Stem Cells Diva ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
A.1 Differential Expression Appendix<br />
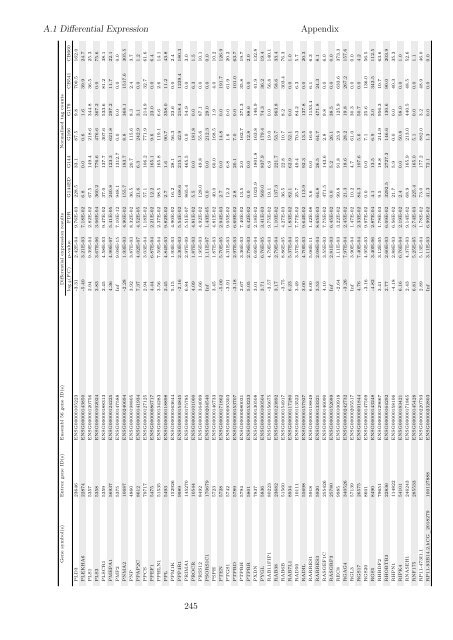
Gene symbol(s) Entrez gene ID(s) Ensembl 56 gene ID(s) Differential expression results Normalised tag counts<br />
log 2(F C) p-value FDR G144ED G144 G166 G179 CB541 CB660<br />
PLD3 23646 ENSG00000105223 -3.31 2.42E-04 9.76E-03 228.5 33.1 87.5 9.8 700.5 162.0<br />
PLEKHA6 22874 ENSG00000143850 -5.40 3.21E-03 7.18E-02 6.8 0.0 0.6 1.6 39.9 24.7<br />
PLS1 5357 ENSG00000120756 2.94 9.39E-04 2.82E-02 67.5 149.4 218.6 344.8 36.5 25.3<br />
PLS3 5358 ENSG00000102024 3.83 3.67E-06 3.06E-04 360.2 776.6 476.6 367.2 0.0 75.6<br />
PLSCR1 5359 ENSG00000188313 2.45 4.58E-03 9.47E-02 37.0 137.7 307.6 533.6 81.2 38.1<br />
PMEPA1 56937 ENSG00000124225 4.36 4.99E-07 5.41E-05 240.8 123.3 621.8 297.2 11.7 22.1<br />
PMP2 5375 ENSG00000147588 Inf 9.08E-15 7.10E-12 948.1 1312.7 0.0 0.0 0.0 0.0<br />
PNMA2 10687 ENSG00000240694 -2.28 1.93E-03 4.90E-02 155.7 183.7 8.8 368.1 1517.6 305.5<br />
PNP 4860 ENSG00000198805 3.92 3.87E-03 8.32E-02 36.5 20.7 115.6 8.3 2.4 3.7<br />
PPAP2C 8612 ENSG00000141934 7.37 4.02E-07 4.52E-05 21.6 6.3 242.9 3.1 0.0 1.2<br />
PPCS 79717 ENSG00000127125 2.94 3.03E-04 1.18E-02 57.1 106.2 771.9 214.9 32.7 61.6<br />
PPEF1 5475 ENSG00000086717 4.44 6.87E-04 2.21E-02 12.2 165.1 9.6 23.0 0.0 6.4<br />
PPHLN1 51535 ENSG00000134283 3.56 7.59E-04 2.37E-02 98.5 165.8 119.1 6.6 2.6 14.1<br />
PPL 5493 ENSG00000118898 2.45 4.84E-03 9.82E-02 2.7 1.6 90.7 358.0 11.2 43.8<br />
PPM1K 152926 ENSG00000163644 5.15 1.93E-03 4.90E-02 16.2 28.1 56.3 23.6 0.0 2.4<br />
PPP4R1 9989 ENSG00000154845 -2.16 2.30E-03 5.54E-02 168.6 215.1 42.9 218.4 1239.4 180.3<br />
PRIMA1 145270 ENSG00000175785 6.84 2.97E-09 5.51E-07 865.4 465.5 0.0 54.9 0.0 3.0<br />
PROCR 10544 ENSG00000101000 4.09 1.87E-03 4.81E-02 5.5 0.0 181.8 0.0 6.3 1.5<br />
PRSS12 8492 ENSG00000164099 3.66 1.95E-03 4.92E-02 128.0 49.8 55.6 87.1 0.0 10.1<br />
PSORS1C1 170679 ENSG00000204540 Inf 1.11E-07 1.43E-05 0.0 0.0 212.3 29.0 0.0 0.0<br />
PSPH 5723 ENSG00000146733 3.45 1.87E-03 4.81E-02 40.0 60.0 168.5 1.9 4.0 10.2<br />
PTEN 5728 ENSG00000171862 -5.00 5.70E-05 2.94E-03 2.7 0.0 14.8 0.0 191.7 126.9<br />
PTGS1 5742 ENSG00000095303 -3.91 4.31E-03 9.05E-02 12.2 6.8 1.6 0.0 61.9 20.2<br />
PTPRD 5789 ENSG00000153707 -3.18 2.97E-03 6.69E-02 2.8 20.1 7.0 0.0 101.0 63.7<br />
PTPRH 5794 ENSG00000080031 2.67 3.36E-03 7.40E-02 13.5 2.0 162.7 371.3 35.8 19.7<br />
PTPRR 5801 ENSG00000153233 5.05 2.78E-03 6.42E-02 0.0 0.0 12.8 88.6 0.0 2.0<br />
PXDN 7837 ENSG00000130508 3.01 6.69E-04 2.18E-02 1025.9 1801.8 163.8 146.9 41.9 132.8<br />
PYGL 5836 ENSG00000100504 3.71 6.76E-05 3.41E-03 569.0 837.9 179.4 74.3 36.2 19.4<br />
RAB11FIP1 80223 ENSG00000156675 -3.57 4.79E-03 9.75E-02 19.1 6.9 19.0 0.0 56.8 140.1<br />
RAB38 23682 ENSG00000123892 3.17 2.79E-04 1.10E-02 137.3 221.7 53.7 963.8 58.6 33.4<br />
RAB6B 51560 ENSG00000154917 -3.75 8.86E-05 4.27E-03 36.5 22.8 10.7 8.2 303.4 76.3<br />
RAB7L1 8934 ENSG00000117280 6.23 5.37E-04 1.83E-02 82.1 62.9 52.1 0.0 0.0 1.0<br />
RAD50 10111 ENSG00000113522 3.49 3.57E-03 7.77E-02 25.7 49.4 70.3 84.2 6.3 5.7<br />
RADIL 55698 ENSG00000157927 3.00 4.70E-03 9.64E-02 113.9 92.3 13.5 137.8 0.0 20.3<br />
RARRES1 5918 ENSG00000118849 6.00 5.06E-11 1.42E-08 5.6 0.0 16.6 1153.4 6.1 6.3<br />
RARRES3 5920 ENSG00000133321 3.53 2.08E-04 8.65E-03 64.8 26.5 64.7 471.8 24.3 8.1<br />
RASGEF1C 255426 ENSG00000146090 4.10 3.55E-03 7.75E-02 471.5 143.6 2.8 9.8 0.0 6.0<br />
RASGRP3 25780 ENSG00000152689 Inf 2.81E-03 6.45E-02 0.0 5.2 26.1 38.5 0.0 0.0<br />
REC8 9985 ENSG00000100918 -2.64 1.11E-03 3.22E-02 99.8 91.8 23.9 125.9 633.6 373.3<br />
RGAG4 340526 ENSG00000242732 -3.26 7.97E-04 2.45E-02 21.3 18.6 28.2 19.8 267.2 157.6<br />
RGL3 57139 ENSG00000205517 Inf 3.90E-04 1.47E-02 10.2 4.7 61.0 30.3 0.0 0.0<br />
RGS17 26575 ENSG00000091844 4.76 7.40E-04 2.33E-02 84.3 107.6 5.6 50.7 0.0 4.2<br />
RGS20 8601 ENSG00000147509 -3.16 4.95E-03 9.97E-02 0.0 0.0 7.1 25.6 136.0 56.5<br />
RGS5 8490 ENSG00000143248 -4.82 3.40E-06 2.87E-04 4.1 13.5 6.9 3.0 343.5 112.5<br />
RHBDF2 79651 ENSG00000129667 3.41 3.12E-05 1.78E-03 9.5 18.8 214.9 956.3 10.7 63.6<br />
RHOBTB3 22836 ENSG00000164292 2.77 2.60E-03 6.09E-02 3292.5 2727.3 146.6 130.6 90.0 203.9<br />
RHPN1 114822 ENSG00000158106 -4.18 2.88E-03 6.56E-02 21.7 5.9 0.0 0.0 40.3 35.3<br />
RIPK4 54101 ENSG00000183421 6.16 6.76E-04 2.19E-02 2.4 0.0 50.8 58.0 0.0 1.0<br />
RNASEH1 246243 ENSG00000171865 2.43 4.37E-03 9.14E-02 68.9 165.9 213.0 440.5 48.5 52.6<br />
RNF175 285533 ENSG00000145428 6.81 5.25E-05 2.74E-03 225.4 165.0 5.1 0.0 0.0 1.1<br />
RP11-473I1.1 ENSG00000220793 2.89 5.13E-04 1.78E-02 73.0 177.2 882.0 3.2 48.9 46.0<br />
RP11-93B14.2,hCG_2018279 100127888 ENSG00000232803 Inf 3.11E-03 6.99E-02 41.3 72.1 1.3 0.0 0.0 0.0<br />
245