4 - Central Institute of Brackishwater Aquaculture
4 - Central Institute of Brackishwater Aquaculture
4 - Central Institute of Brackishwater Aquaculture
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Natlonal Workshop-cum-Training on Bldnfonnatics and Information Management in <strong>Aquaculture</strong><br />
favoured reglons<br />
275 94.2%<br />
Res~dues in addlt~onal allowed regens<br />
I<br />
I<br />
I<br />
I<br />
------'<br />
Resldues In disallowed reglons<br />
1 1 0.3%<br />
---- .........................................<br />
...............................<br />
I<br />
Number <strong>of</strong> non-glyc~ne and non-prol~ne<br />
res~dues 292 100.0%<br />
AII kgree, Pro) 2<br />
Number <strong>of</strong> glyclne res~dues (shown as<br />
triangles) 30<br />
Number <strong>of</strong> prol~ne res~dues<br />
..............................................<br />
...............................<br />
Total number <strong>of</strong> res~dues<br />
337<br />
..............................................<br />
...............................<br />
M/c bond lengths: 99.5%<br />
0.5% h~ghl~ghted<br />
M/c bond angles: 93.2%<br />
1 1 6.E0/0 h~ghl~ghted<br />
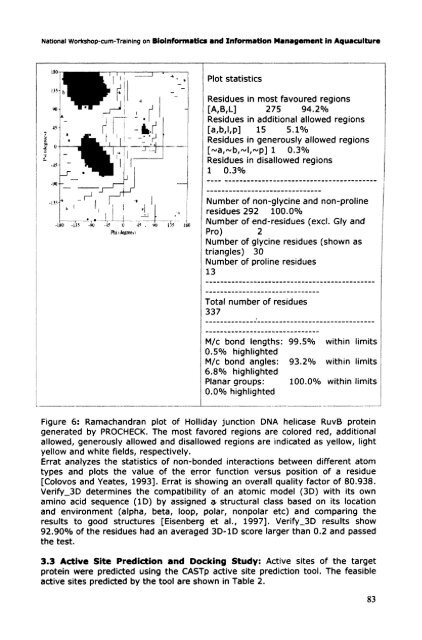
Figure 6: Ramachandran plot <strong>of</strong> Holliday junction DNA helicase RuvB protein<br />
generated by PROCHECK. The most favored regions are colored red, additional<br />
allowed, generously allowed and disallowed regions are indicated as yellow, light<br />
yellow and white fields, respectively.<br />
Errat analyzes the statistics <strong>of</strong> non-bonded interactions between different atom<br />
types and plots the value <strong>of</strong> the error function versus position <strong>of</strong> a residue<br />
[Colovos and Yeates, 19931. Errat is showing an overall quality factor <strong>of</strong> 80.938.<br />
Verify-3D determines the compatibility <strong>of</strong> an atomic model (3D) with its own<br />
amino acid sequence (ID) by assigned & structural class based on its location<br />
and environment (alpha, beta, loop, polar, nonpolar etc) and comparing the<br />
results to good structures [Eisenberg et al., 19971. Verify-3D results show<br />
92.90% <strong>of</strong> the residues had an averaged 3D-1D score larger than 0.2 and passed<br />
the test.<br />
3.3 Active Site Prediction and Docking Study: Active sites <strong>of</strong> the target<br />
protein were predicted using the CASTp active site prediction tool. The feasible<br />
active sites predicted by the tool are shown in Table 2.<br />
I<br />
I<br />
wlthln l~m~ts<br />
I Planar groups: 100.0% wlth~n llmlts<br />
0.0% h~ghl~ghted<br />
I