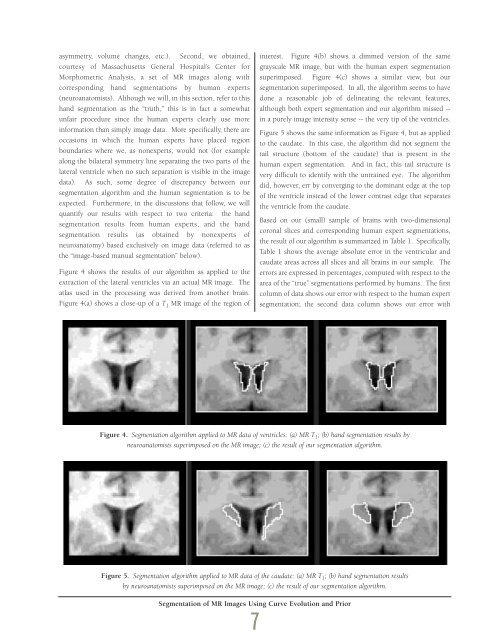
asymmetry, volume changes, etc.). Second, we obtained,courtesy of Massachusetts General Hospital’s Center forMorphometric Analysis, a set of MR images along withcorresponding hand segmentations by human experts(neuroanatomists). Although we will, in this section, refer to thishand segmentation as the “truth,” this is in fact a somewhatunfair procedure since the human experts clearly use moreinformation than simply image data. More specifically, there areoccasions in which the human experts have placed regionboundaries where we, as nonexperts, would not (for examplealong the bilateral symmetry line separating the two parts of thelateral ventricle when no such separation is visible in the imagedata). As such, some degree of discrepancy between oursegmentation algorithm and the human segmentation is to beexpected. Furthermore, in the discussions that follow, we willquantify our results with respect to two criteria: the handsegmentation results from human experts, and the handsegmentation results (as obtained by nonexperts ofneuroanatomy) based exclusively on image data (referred to asthe “image-based manual segmentation” below).Figure 4 shows the results of our algorithm as applied to theextraction of the lateral ventricles via an actual MR image. Theatlas used in the processing was derived from another brain.Figure 4(a) shows a close-up of a T 1 MR image of the region ofinterest. Figure 4(b) shows a dimmed version of the samegrayscale MR image, but with the human expert segmentationsuperimposed. Figure 4(c) shows a similar view, but oursegmentation superimposed. In all, the algorithm seems to havedone a reasonable job of delineating the relevant features,although both expert segmentation and our algorithm missed --in a purely image intensity sense -- the very tip of the ventricles.Figure 5 shows the same information as Figure 4, but as appliedto the caudate. In this case, the algorithm did not segment thetail structure (bottom of the caudate) that is present in thehuman expert segmentation. And in fact, this tail structure isvery difficult to identify with the untrained eye. The algorithmdid, however, err by converging to the dominant edge at the topof the ventricle instead of the lower contrast edge that separatesthe ventricle from the caudate.Based on our (small) sample of brains with two-dimensionalcoronal slices and corresponding human expert segmentations,the result of our algorithm is summarized in Table 1. Specifically,Table 1 shows the average absolute error in the ventricular andcaudate areas across all slices and all brains in our sample. Theerrors are expressed in percentages, computed with respect to thearea of the “true” segmentations performed by humans. The firstcolumn of data shows our error with respect to the human expertsegmentation; the second data column shows our error withFigure 4. Segmentation algorithm applied to MR data of ventricles: (a) MR T 1 ; (b) hand segmentation results byneuroanatomists superimposed on the MR image; (c) the result of our segmentation algorithm.Figure 5. Segmentation algorithm applied to MR data of the caudate: (a) MR T 1 ; (b) hand segmentation resultsby neuroanatomists superimposed on the MR image; (c) the result of our segmentation algorithm.Segmentation of MR Images Using Curve Evolution and Prior7
Table 1. A summary of segmentation algorithm error with respectto human segmentation.respect to an independent image-based manual segmentation bysomeone with knowledge only of image processing and not ofneuroanatomy.As Table 1, shows, there is a significant difference in performancewhen comparing the algorithm to human expert segmentationversus an image-only-based human segmentation. Aperformance of 20-percent error as compared to human experts,while not great, was also not unexpected given the extent towhich such segmentations deviated from image data. A 5-percent error as compared to nonexpert segmentation, however,reassured us that the algorithm is properly exploiting the imagefeatures as well as the prior information in deriving the resultingsegmentations.Concluding RemarksIn summary, we have presented a method for segmentinganatomical structures using only minimal operator interactions.This method achieves good performance when compared tohuman segmentations based solely on image intensities. Theproposed formulation can be a powerful new tool in assistingscientists and clinicians in evaluating patient ailments andmaking volumetric measurements. This formulation continuesthe work of numerous researchers in the field of variationalimage processing and uses a new formulation recently describedby Shah (Ref. [12]) for combining into a single functional theability to perform nonlinear diffusion, estimate an edge strengthfunction, perform curve evolution, and ultimately obtain asegmentation. When applied to the problems of delineating thelateral ventricles and caudate, the formulation yieldswell-behaved curves that capture the essence of the structures ofinterest, while remaining robust with respect to the uneven biasfield effects, low-contrast edges, and even shape distortions. Thistool is sufficiently general to assist the human operator in avariety of scenarios, including segmentation from a predefinedatlas, as well as segmentation of a structure of interest using anearlier segmentation as the reference. In particular, although wehave demonstrated our approach using a predefined atlas, ourempirical tests have shown great robustness when segmentingone slice of MR data by bootstrapping from the segmentation ofthe previous slice.Two significant problems still persist in our current formulation.First, the method is computationally intensive. Even when theimage is limited to the region of interest (e.g., the areacontaining the lateral ventricles and caudate), it takes severalminutes to estimate the edge strength function as well as performthe curve evolution using a Sun Sparc20 workstation. Ideally, wewould like to achieve near real time so that the clinician canreceive immediate quantitative results. Second, at present, themethod does not perform well in sharp corners due to the factthat curve evolution evolves with a velocity proportional tocurvature. This problem is particularly acute near the horns ofthe lateral ventricles where the corners can be quite sharp. Weare currently working on a remedy for this problem.AcknowledgmentWe gratefully acknowledge Drs. Kennedy and Worth ofMassachusetts General Hospital for providing the MR imagery.We are also indebted to Anthony Sacramone for his able support,and to David Suh for his programming support on the thin-platewarping algorithm.References[1] Filipek, P., C. Richelme, D. Kennedy, and V. Caviness, “TheYoung Adult Human Brain: an MRI-Based MorphometricAnalysis,” Cereb. Cortex, 4(4), July-August 1994.[2] Giedd, F., J. Snell, N. Lange, J. Rajapakse, B. Casey, P. Kozuch,A. Vaituzis, Y. Vauss, S. Hamburger, D. Kaysen, and J.Raporport, “Quantitative Magnetic Resonance Imaging ofHuman Brain Development: Ages 4 - 18,” Cereb Cortex, 6(4),July-August 1996.[3] Jernigan, T., G. Press, and J. Hesselink, “Methods forMeasuring Brain Morphologic Features on MagneticResonance Images,” Arch. Neurology, 47, January 1990.[4] Sullivan, E., L. Marsh, D. Mathalon, K. Lim, and A.Pfefferbaum, “Age-Related Decline in MRI Volumes ofTemporal Lobe Gray Matter But Not Hippocamus,”Neurobiol. Aging, 16(4), July-August 1995.[5] Golden, N., M. Ashtari, M. Kohn, M. Patel, M. Jacobson, A.Fletcher, and I. Shenker, “Reversibility of Cerebral VentricularEnlargement in Anorexia Nervosa, Demonstrated byQuantitative Magnetic Resonance Imaging,” J. Pediatrics,128(2), February 1996.[6] Castellanos, F., J. Giedd, W. Marsh, S. Hamburger, A. Vaituzis,D. Dickstein, S. Sarfatti, Y. Vauss, J. Snell, N. Lange, D.Segmentation of MR Images Using Curve Evolution and Prior8
- Page 2 and 3: Letter from thePresident and CEO,Vi
- Page 4 and 5: Information TechnologyMilton AdamsE
- Page 6 and 7: BiographyMilton Adams has been at D
- Page 9 and 10: Figure 1 represents a functional de
- Page 11 and 12: Programs. In effect, these controll
- Page 13 and 14: Although the terminal area traffic
- Page 15 and 16: Table 2. ATFM performance evaluatio
- Page 17 and 18: In the experiments, a nominal capac
- Page 19 and 20: [3] Wambsganss, Michael C. “Colla
- Page 21 and 22: Guidance, Navigation, and Control A
- Page 23 and 24: A Control Lyapunov FunctionApproach
- Page 25 and 26: x( 0) ∈ X and w(t) ∈Wfor all t
- Page 27 and 28: (b) Select a quadratic RCLF V i (x)
- Page 29 and 30: at each grid point. In the case w 1
- Page 31 and 32: References[1] Ball, J.A. and A.J. v
- Page 33 and 34: Guidance, Navigation, and Control A
- Page 35 and 36: Relative and Differential GPSData T
- Page 37 and 38: The first term on the right in the
- Page 39 and 40: H R# δρ R,GPS -H A# δρ A,GPSThi
- Page 41 and 42: selection; and (3) shown that the a
- Page 43 and 44: Guidance, Navigation, and Control A
- Page 45 and 46: Segmentation of MR ImagesUsing Curv
- Page 47 and 48: (3)where ν now represents a contin
- Page 49: Experimental ResultsThe results of
- Page 53 and 54: Guidance, Navigation,and ControlJim
- Page 55 and 56: BiographyGeorge SchmidtGeorge Schmi
- Page 57 and 58: clock and ephemeris errors, as well
- Page 59 and 60: maintained in a rigid structure, wh
- Page 61 and 62: Table 5. “Typical” absolute GPS
- Page 63 and 64: performed, then the target location
- Page 65 and 66: tightly-coupled system, however, ca
- Page 67 and 68: Concluding RemarksRecent progress i
- Page 69 and 70: As real-time systems evolve into th
- Page 71 and 72: Advanced Fault-TolerantComputing fo
- Page 73 and 74: The Viking and Voyager were both in
- Page 75 and 76: Containment Regions (FCRs). There a
- Page 77 and 78: well as reversing the whole process
- Page 79 and 80: As real-time systems evolve into th
- Page 81 and 82: Automated Station-Keepingfor Satell
- Page 83 and 84: Figure 2. Minimum elevation angles
- Page 85 and 86: anomaly M and/or the ascending node
- Page 87 and 88: However, since optimization and rec
- Page 89 and 90: is maintained in the Northern Hemis
- Page 91 and 92: autonomy. It must have the ability
- Page 93 and 94: [31] Neelon, Joseph G., Jr., Paul J
- Page 95 and 96: Draper’s primary goal is to Drape
- Page 97 and 98: )Rotordynamic Modelingof an Activel
- Page 99 and 100: Eq. (9) becomes:λ[ R ] { Φ } = [
- Page 101 and 102:
chosen to be 24, for a total of 48
- Page 103 and 104:
InertialInstruments/MechanicalDesig
- Page 105 and 106:
BiographyJeffrey Borenstein is curr
- Page 107 and 108:
process step. Process information i
- Page 109 and 110:
Figure 4. Control chart for boron d
- Page 111 and 112:
References[1] Barbour, N., J. Conne
- Page 113 and 114:
Draper Laboratory continues to engi
- Page 115 and 116:
Validating the Validating Tool:Defi
- Page 117 and 118:
calculates miscellaneous terms, suc
- Page 119 and 120:
Table 1. Suggested specification sh
- Page 121 and 122:
User Accuracy as aFunction of Simul
- Page 123 and 124:
20-min averaging, this clock lockin
- Page 125 and 126:
Table 2. Sample high-level summary
- Page 127 and 128:
AcknowledgmentR.L. Greenspan, J.A.
- Page 129 and 130:
Systems IntegrationRich MartoranaPe
- Page 131 and 132:
BiographyAnthony Kourepenis is an A
- Page 133 and 134:
control is employed to maintain the
- Page 135 and 136:
Table 1. Summary of automotive yaw
- Page 137 and 138:
Resolution (60 Hz) deg/h10000000100
- Page 139 and 140:
References[1] Greiff, P., B. Boxenh
- Page 141 and 142:
Guidance, Navigation, and Control A
- Page 143 and 144:
An Integrated Safety AnalysisMethod
- Page 145 and 146:
Infrastructure ModelsSystemRequirem
- Page 147 and 148:
Figures 6 and 7 illustrate the bloc
- Page 149 and 150:
Notice that each flight track descr
- Page 151 and 152:
Table 7. Safety statistics at 1700-
- Page 153 and 154:
Guidance, Navigation, and Control A
- Page 155 and 156:
An Optimal Guidance Law forPlanetar
- Page 157 and 158:
Note that the states in the three d
- Page 159 and 160:
Crossrange (Kft)10090807060504030Cl
- Page 161 and 162:
The 1997 Charles StarkDraper PrizeT
- Page 163 and 164:
The 1997 Charles StarkDraper Prize1
- Page 165 and 166:
“Draper encourages its personnel
- Page 167 and 168:
Gimballed Vibrating GyroscopeHaving
- Page 169 and 170:
“Draper encourages its personnel
- Page 171 and 172:
Optical Source Isolator withPolariz
- Page 173 and 174:
“Draper encourages its personnel
- Page 175 and 176:
Hunting Suppressor forPolyphase Ele
- Page 177 and 178:
“Draper encourages its personnel
- Page 179 and 180:
Sensor Having an Off-Frequency Driv
- Page 181 and 182:
proof mass from transients and enha
- Page 183 and 184:
1997 Published PapersThe following
- Page 185 and 186:
monitoring of space structures and
- Page 187 and 188:
measured by kinematic degrees of fr
- Page 189 and 190:
i.e., what percent of the earth’s
- Page 191 and 192:
McConley, M. W.; Dahleh, M. A.; Fer
- Page 193 and 194:
unaffordable, or even misguided. Bu
- Page 195 and 196:
The Draper DistinguishedPerformance
- Page 197:
Educational Activitiesat Draper Lab