- Page 2:
Advanced Techniques in Diagnostic M
- Page 6:
Yi-Wei Tang Molecular Infectious Di
- Page 10:
vi Contributors Wonder Drake Depart
- Page 14:
viii Contributors Jacques Schrenzel
- Page 18:
Preface Clinical microbiologists ar
- Page 22:
Contents Part I Techniques 1 Automa
- Page 26:
Contents xv 22 Review of Molecular
- Page 30:
1 Automated Blood Cultures XIANG Y.
- Page 34:
Interpretation of Significance 1. A
- Page 38:
1. Automated Blood Cultures 7 conta
- Page 42:
1. Automated Blood Cultures 9 Crump
- Page 46:
2 Urea Breath Tests for Detection o
- Page 50:
2. Urea Breath Tests 13 and point o
- Page 54:
2. Urea Breath Tests 15 Other detec
- Page 58:
TABLE 2.1. Comparison of 13 C-urea
- Page 62:
2. Urea Breath Tests 19 Bazzoli, F.
- Page 66:
2. Urea Breath Tests 21 Logan, R. (
- Page 70:
3 Rapid Antigen Tests SHELDON CAMPB
- Page 74:
3. Rapid Antigen Tests 25 antigen i
- Page 78:
3. Rapid Antigen Tests 27 either on
- Page 82:
3. Rapid Antigen Tests 29 and moves
- Page 86:
3. Rapid Antigen Tests 31 Applicati
- Page 90:
Fungi Cryptococcus Agglutination, E
- Page 94:
Adenovirus, enteric types 40,41 EIA
- Page 98:
3. Rapid Antigen Tests 37 Antigen t
- Page 102:
3. Rapid Antigen Tests 39 retains a
- Page 106:
3. Rapid Antigen Tests 41 Wilhelmi,
- Page 110:
4. Advanced Antibody Detection 43 D
- Page 114:
TABLE 4.1. Types of antibody detect
- Page 118:
4. Advanced Antibody Detection 47 c
- Page 122:
4. Advanced Antibody Detection 49 U
- Page 126:
4. Advanced Antibody Detection 51 a
- Page 130:
4. Advanced Antibody Detection 53 a
- Page 134:
4. Advanced Antibody Detection 55 H
- Page 138:
4. Advanced Antibody Detection 57 r
- Page 142:
4. Advanced Antibody Detection 59 A
- Page 146:
4. Advanced Antibody Detection 61 M
- Page 150:
5 Phenotypic Testing of Bacterial A
- Page 154:
5. Antimicrobial Susceptibility Tes
- Page 158:
5. Antimicrobial Susceptibility Tes
- Page 162:
5. Antimicrobial Susceptibility Tes
- Page 166:
Susceptibility Tests for Fastidious
- Page 170:
5. Antimicrobial Susceptibility Tes
- Page 174:
5. Antimicrobial Susceptibility Tes
- Page 178:
5. Antimicrobial Susceptibility Tes
- Page 182:
References 5. Antimicrobial Suscept
- Page 186:
5. Antimicrobial Susceptibility Tes
- Page 190:
5. Antimicrobial Susceptibility Tes
- Page 194:
6. Biochemical Profile-Based Microb
- Page 198:
6. Biochemical Profile-Based Microb
- Page 202:
6. Biochemical Profile-Based Microb
- Page 206:
6. Biochemical Profile-Based Microb
- Page 210:
6. Biochemical Profile-Based Microb
- Page 214:
6. Biochemical Profile-Based Microb
- Page 218:
6. Biochemical Profile-Based Microb
- Page 222:
6. Biochemical Profile-Based Microb
- Page 226:
6. Biochemical Profile-Based Microb
- Page 230:
6. Biochemical Profile-Based Microb
- Page 234:
6. Biochemical Profile-Based Microb
- Page 238:
6. Biochemical Profile-Based Microb
- Page 242:
6. Biochemical Profile-Based Microb
- Page 246:
6. Biochemical Profile-Based Microb
- Page 250:
6. Biochemical Profile-Based Microb
- Page 254:
6. Biochemical Profile-Based Microb
- Page 258:
7 Rapid Bacterial Characterization
- Page 262:
7. MALDI-TOF MS 119 photons followe
- Page 266:
7. MALDI-TOF MS 121 for analysis by
- Page 270:
7. MALDI-TOF MS 123 Sample wells Ca
- Page 274:
7. MALDI-TOF MS 125 also given. Thi
- Page 278:
7. MALDI-TOF MS 127 characteristics
- Page 282:
References 7. MALDI-TOF MS 129 Ande
- Page 286:
7. MALDI-TOF MS 131 Hindre, T., Did
- Page 290:
7. MALDI-TOF MS 133 von Wintzingero
- Page 294:
8. Probe-Based Microbial Detection
- Page 298:
8. Probe-Based Microbial Detection
- Page 302:
8. Probe-Based Microbial Detection
- Page 306:
8. Probe-Based Microbial Detection
- Page 310:
9 Pulsed-Field Gel Electrophoresis
- Page 314:
9. Pulsed-Field Gel Electrophoresis
- Page 318:
9. Pulsed-Field Gel Electrophoresis
- Page 322:
PFGE Performance Characteristics 9.
- Page 326:
9. Pulsed-Field Gel Electrophoresis
- Page 330:
9. Pulsed-Field Gel Electrophoresis
- Page 334:
9. Pulsed-Field Gel Electrophoresis
- Page 338:
9. Pulsed-Field Gel Electrophoresis
- Page 342:
TABLE 10.1. Nucleic acid amplificat
- Page 346:
10. In Vitro Nucleic Acid Amplifica
- Page 350:
10. In Vitro Nucleic Acid Amplifica
- Page 354:
10. In Vitro Nucleic Acid Amplifica
- Page 358:
11. PCR and Its Variations 167 Prin
- Page 362:
11. PCR and Its Variations 169 Exte
- Page 366:
11. PCR and Its Variations 171 targ
- Page 370:
11. PCR and Its Variations 173 bind
- Page 374:
11. PCR and Its Variations 175 Reve
- Page 378:
11. PCR and Its Variations 177 ampl
- Page 382:
11. PCR and Its Variations 179 pres
- Page 386:
11. PCR and Its Variations 181 Soft
- Page 390:
11. PCR and Its Variations 183 Saik
- Page 394: 12. Non-PCR Amplification 185 1989;
- Page 398: 12. Non-PCR Amplification 187 FIGUR
- Page 402: 12. Non-PCR Amplification 189 This
- Page 406: 12. Non-PCR Amplification 191 FIGUR
- Page 410: 12. Non-PCR Amplification 193 and c
- Page 414: 12. Non-PCR Amplification 195 used
- Page 418: 12. Non-PCR Amplification 197 do no
- Page 422: Application of the SDA Technique Re
- Page 426: 12. Non-PCR Amplification 201 Ancho
- Page 430: 12. Non-PCR Amplification 203 Baner
- Page 434: 12. Non-PCR Amplification 205 Guilf
- Page 438: 12. Non-PCR Amplification 207 Nilss
- Page 442: 12. Non-PCR Amplification 209 Wang,
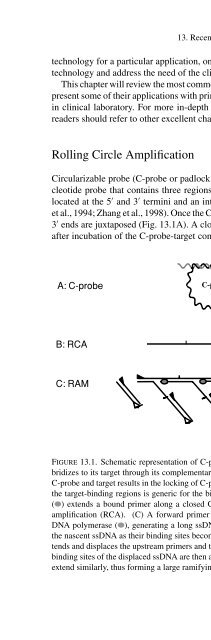
- Page 448: 212 D. Zhang et al. TABLE 13.1. Com
- Page 452: 214 D. Zhang et al. scanner after h
- Page 456: 216 D. Zhang et al. FIGURE 13.3. Sc
- Page 460: 218 D. Zhang et al. FIGURE 13.4. Sc
- Page 464: 220 D. Zhang et al. A. B. Invasive
- Page 468: 222 D. Zhang et al. FIGURE 13.6. Pr
- Page 472: 224 D. Zhang et al. The mecA probe
- Page 476: 226 D. Zhang et al. Landegren, U.,
- Page 480: 14 Signal Amplification Techniques:
- Page 484: 230 Y. F. Wang Virus (3)* (2) (4)*
- Page 488: 232 Y. F. Wang 1. Denaturation 2. H
- Page 492: 234 Y. F. Wang Application of the T
- Page 496:
236 Y. F. Wang specimen. A study (K
- Page 500:
238 Y. F. Wang amplification techno
- Page 504:
240 Y. F. Wang Cuzick, J., Szarewsk
- Page 508:
242 Y. F. Wang Qian, X., & Lloyd, R
- Page 512:
244 R. P. Podzorski, M. Loeffelholz
- Page 516:
246 R. P. Podzorski, M. Loeffelholz
- Page 520:
248 R. P. Podzorski, M. Loeffelholz
- Page 524:
250 R. P. Podzorski, M. Loeffelholz
- Page 528:
252 R. P. Podzorski, M. Loeffelholz
- Page 532:
254 R. P. Podzorski, M. Loeffelholz
- Page 536:
256 R. P. Podzorski, M. Loeffelholz
- Page 540:
258 R. P. Podzorski, M. Loeffelholz
- Page 544:
260 R. P. Podzorski, M. Loeffelholz
- Page 548:
262 R. P. Podzorski, M. Loeffelholz
- Page 552:
16 Direct Nucleotide Sequencing for
- Page 556:
266 Tao Hong approximately 10 min (
- Page 560:
268 Tao Hong frequently caused prob
- Page 564:
270 Tao Hong for the use of the 16S
- Page 568:
272 Tao Hong of MRSA. The coagulase
- Page 572:
274 Tao Hong Gürtler, V., & Barrie
- Page 576:
17 Microarray-Based Microbial Ident
- Page 580:
278 T.J. Gentry and J. Zhou These a
- Page 584:
280 T.J. Gentry and J. Zhou the des
- Page 588:
282 T.J. Gentry and J. Zhou Communi
- Page 592:
284 T.J. Gentry and J. Zhou PCR-bas
- Page 596:
286 T.J. Gentry and J. Zhou Althoug
- Page 600:
288 T.J. Gentry and J. Zhou Korczak
- Page 604:
290 T.J. Gentry and J. Zhou Willse,
- Page 608:
292 X. Qin allow real-time monitori
- Page 612:
294 X. Qin which the observed fluor
- Page 616:
296 X. Qin that there would be no G
- Page 620:
298 X. Qin 2005). Furthermore, the
- Page 624:
300 X. Qin Cattoli, G., Drago, A.,
- Page 628:
302 X. Qin Kostrikis, L.G., Touloum
- Page 632:
304 X. Qin Ririe, K.M., Rasmussen,
- Page 636:
19 Amplification Product Inactivati
- Page 640:
308 S. Sefers and Y-W. Tang Most de
- Page 644:
310 S. Sefers and Y-W. Tang UNG Pro
- Page 648:
312 S. Sefers and Y-W. Tang 4' Psor
- Page 652:
314 S. Sefers and Y-W. Tang Isopsor
- Page 656:
316 S. Sefers and Y-W. Tang used is
- Page 660:
318 S. Sefers and Y-W. Tang Conclus
- Page 664:
Part II Applications
- Page 668:
324 X.Y. Han TABLE 20.1. The number
- Page 672:
326 X.Y. Han Homology Search and Re
- Page 676:
328 X.Y. Han TABLE 20.2. Examples o
- Page 680:
330 X.Y. Han Conclusion In summary,
- Page 684:
332 X.Y. Han Shimizu, T., Ohtani, K
- Page 688:
TABLE 21.1. Licensed / Approved Cli
- Page 692:
TABLE 21.1. (Continued) Tradename(s
- Page 696:
TABLE 21.1. (Continued) Tradename(s
- Page 700:
340 Y. Hu and I. Hirshfield gel ele
- Page 704:
342 Y. Hu and I. Hirshfield Since 1
- Page 708:
344 Y. Hu and I. Hirshfield they ar
- Page 712:
346 Y. Hu and I. Hirshfield Antibod
- Page 716:
348 Y. Hu and I. Hirshfield all ove
- Page 720:
350 Y. Hu and I. Hirshfield chances
- Page 724:
352 Y. Hu and I. Hirshfield Tang, Y
- Page 728:
354 A. C. T. Lo and K. M. Kam in tu
- Page 732:
356 A. C. T. Lo and K. M. Kam is no
- Page 736:
358 A. C. T. Lo and K. M. Kam stabl
- Page 740:
360 A. C. T. Lo and K. M. Kam Stran
- Page 744:
362 A. C. T. Lo and K. M. Kam organ
- Page 748:
364 A. C. T. Lo and K. M. Kam probe
- Page 752:
366 A. C. T. Lo and K. M. Kam major
- Page 756:
368 A. C. T. Lo and K. M. Kam Molec
- Page 760:
370 A. C. T. Lo and K. M. Kam a sec
- Page 764:
372 A. C. T. Lo and K. M. Kam cervi
- Page 768:
374 A. C. T. Lo and K. M. Kam E6 an
- Page 772:
376 A. C. T. Lo and K. M. Kam M. (2
- Page 776:
378 A. C. T. Lo and K. M. Kam Hippe
- Page 780:
380 A. C. T. Lo and K. M. Kam Larse
- Page 784:
382 A. C. T. Lo and K. M. Kam Nobbe
- Page 788:
384 A. C. T. Lo and K. M. Kam chlam
- Page 792:
386 A. C. T. Lo and K. M. Kam ureal
- Page 796:
388 A. Kilic and W. Drake Lipids wi
- Page 800:
390 A. Kilic and W. Drake America,
- Page 804:
392 A. Kilic and W. Drake MTB from
- Page 808:
394 A. Kilic and W. Drake with anti
- Page 812:
396 A. Kilic and W. Drake Molecular
- Page 816:
398 A. Kilic and W. Drake and monit
- Page 820:
400 A. Kilic and W. Drake system; t
- Page 824:
402 A. Kilic and W. Drake transillu
- Page 828:
404 A. Kilic and W. Drake Real-Time
- Page 832:
406 A. Kilic and W. Drake Caws, M.,
- Page 836:
408 A. Kilic and W. Drake Marttila,
- Page 840:
410 A. Kilic and W. Drake Torres, M
- Page 844:
412 P. Francois and J. Schrenzel an
- Page 848:
414 P. Francois and J. Schrenzel th
- Page 852:
416 P. Francois and J. Schrenzel A
- Page 856:
418 P. Francois and J. Schrenzel St
- Page 860:
420 P. Francois and J. Schrenzel Ac
- Page 864:
422 P. Francois and J. Schrenzel or
- Page 868:
424 P. Francois and J. Schrenzel Lo
- Page 872:
426 P. Francois and J. Schrenzel Va
- Page 876:
428 D. Ernst et al. several microme
- Page 880:
430 D. Ernst et al. reanalysis to d
- Page 884:
432 D. Ernst et al. Neisseria menin
- Page 888:
434 D. Ernst et al. A. B. SEB-beads
- Page 892:
436 D. Ernst et al. IL-6, IL-8, and
- Page 896:
pg/ml 120 100 80 60 40 20 0 120 100
- Page 900:
440 D. Ernst et al. Although interf
- Page 904:
442 D. Ernst et al. Lal, G., Balmer
- Page 908:
26 Molecular Strain Typing Using Re
- Page 912:
446 S. R. Frye and M. Healy FIGURE
- Page 916:
448 S. R. Frye and M. Healy Automat
- Page 920:
450 S. R. Frye and M. Healy TABLE 2
- Page 924:
452 S. R. Frye and M. Healy Noncomm
- Page 928:
454 S. R. Frye and M. Healy 2001).
- Page 932:
456 S. R. Frye and M. Healy (Stempe
- Page 936:
458 S. R. Frye and M. Healy isolate
- Page 940:
460 S. R. Frye and M. Healy molecul
- Page 944:
462 S. R. Frye and M. Healy Boerlin
- Page 948:
464 S. R. Frye and M. Healy Fey, P.
- Page 952:
466 S. R. Frye and M. Healy Kwara,
- Page 956:
468 S. R. Frye and M. Healy Pfaller
- Page 960:
470 S. R. Frye and M. Healy Tang, Y
- Page 964:
27 Molecular Differential Diagnoses
- Page 968:
474 J. Han The technology advanceme
- Page 972:
476 J. Han Targets Optimal Conditio
- Page 976:
478 J. Han TABLE 27.1. Comparison o
- Page 980:
480 J. Han TABLE 27.3. Comparison o
- Page 984:
482 J. Han suspension hybridization
- Page 988:
484 J. Han fluorescent dye is remov
- Page 992:
486 J. Han TABLE 27.5. List of path
- Page 996:
TABLE 27.8. Detection results of th
- Page 1000:
490 J. Han common in HAI. In additi
- Page 1004:
492 J. Han TABLE 27.12. Detection r
- Page 1008:
494 J. Han TABLE 27.14. List of pat
- Page 1012:
496 J. Han TABLE 27.18. Detectable
- Page 1016:
TABLE 27.20. List of gene targets i
- Page 1020:
500 J. Han Benefits and Impact: Del
- Page 1024:
502 J. Han significantly increasing
- Page 1028:
504 J. Han Hindiyeh, M., Hillyard D
- Page 1032:
506 E. M. Marlowe and D. M. Wolk Al
- Page 1036:
TABLE 28.1. Automated specimen proc
- Page 1040:
510 E. M. Marlowe and D. M. Wolk Au
- Page 1044:
512 E. M. Marlowe and D. M. Wolk La
- Page 1048:
514 E. M. Marlowe and D. M. Wolk me
- Page 1052:
516 E. M. Marlowe and D. M. Wolk mo
- Page 1056:
518 E. M. Marlowe and D. M. Wolk Mo
- Page 1060:
520 E. M. Marlowe and D. M. Wolk Ha
- Page 1064:
522 E. M. Marlowe and D. M. Wolk Sa
- Page 1068:
Index A acetate utilization, 100 Ac
- Page 1072:
Index 527 rapid antigen tests, 27-2
- Page 1076:
Index 529 flow cytometric immunoass
- Page 1080:
Index 531 Human T-Lymphotropic Viru
- Page 1084:
Index 533 MLEE. See multilocus enzy
- Page 1088:
Index 535 PNA. See peptide-nucleic
- Page 1092:
Index 537 Roseomonas, 328-329 rotav
- Page 1096:
Index 539 T TaqMan probes, 293-296