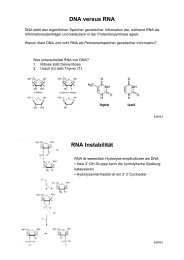
ZMBH J.Bericht 2000 - Zentrum für Molekulare Biologie der ...
ZMBH J.Bericht 2000 - Zentrum für Molekulare Biologie der ...
ZMBH J.Bericht 2000 - Zentrum für Molekulare Biologie der ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
genes of the procyclic form of Trypanosoma brucei.<br />
Mol. Biochem. Parasit. 103, 99-100.<br />
Clayton, C.E., Maier, A., Lorenz, P., Blattner, J., Helfert,<br />
S., Krieger, S. (<strong>2000</strong>). Strange characteristics of<br />
kinetoplastid protists: targets for antiparasitic chemotherapy?<br />
Nova Acta Leopoldina 80, 15-25.<br />
Clayton, C.E. (1999). Why do we need standard<br />
genetic nomenclature for parasites? Acta Tropica (in<br />
press)<br />
THESES<br />
Diploma<br />
Ding, M. (1998). Charakterisierung von Organellen in<br />
Toxoplasma gondii.<br />
Herberger, M. (1999). Klonierung und Expression<br />
eines Cysteine Rich Acidic Integral Membrane<br />
(CRAM)-GFP-Fusionsgens in Trypanosoma brucei<br />
brucei.<br />
Dissertations<br />
Krieger, S. (1998). Die Trypanothionreduktase bei<br />
Trypanosoma brucei:Etablierung und Chatakterisie–<br />
rung einer TR-defizienten Zelllinie.<br />
Maier, A. (1999). Funktionelle Charakterisierung<br />
glykosomaler Membranproteine aus Trypanosoma<br />
brucei.<br />
60<br />
STRUCTURE OF THE GROUP<br />
E-mail: clayton@zmbh.uni-heidelberg.de<br />
Group lea<strong>der</strong>: Clayton, Christine, Prof. Dr.<br />
Postdoctoral fellows<br />
Ansorge, Iris, Dr.<br />
Estévez, Antonio, PhD*<br />
Quilada, Luis, PhD*<br />
Voncken, Frank, PhD*<br />
Ph.D. students Ding, Martine, Dipl. Biol.*<br />
Drodzd, Maciej, Dipl. Biol.<br />
Guerra-Giraldez, Christina, MSc.<br />
Helfert, Sandra, Dipl. Biol.<br />
Irmer, Henriette, Dipl. Biol.<br />
Krieger, Stephan, Dipl. Biol.<br />
Maier, Alexan<strong>der</strong>, Dipl. Biol.<br />
Diploma students Ding, Martina *<br />
Schulreich, Sebastian *<br />
Herberger, Michael *<br />
Techn. assistant Hartmann, Claudia<br />
*part of the time reported<br />
Bernhard Dobberstein<br />
Protein Targeting and Intracellular<br />
Sorting<br />
Protein translocation across the membrane of the endoplasmic<br />
reticulum (ER) involves cytosolic chaperones,<br />
docking receptors, a translocation channel (translocon)<br />
and in some cases a “translocation motor” which<br />
drives the actual translocation (for review see Schatz<br />
and Dobberstein, 1996). Once in the ER, proteins are<br />
folded, modified and - after a quality control - packed<br />
into vesicles and transported to the Golgi complex and<br />
the trans-Golgi network. From there they can either<br />
be further transported to the plasma membrane or to<br />
organelles of the endosomal system. A major focus of<br />
the work of our group is the analysis of mechanisms<br />
involved in targeting proteins to the ER membrane<br />
and in their translocation across or insertion into this<br />
membrane. Special emphasis is on the control and<br />
regulation of these processes.<br />
Cotranslational targeting of nascent secretory and<br />
membrane proteins to the ER membrane involves<br />
the signal recognition particle (SRP) and its receptor<br />
(SRP-receptor or docking protein). SRP interacts with<br />
the signal sequence of the nascent proteins and mediates<br />
their transfer to the ER membrane. The SRP<br />
receptor catalyzes the release of the nascent chain<br />
from SRP and its insertion into the translocon. SRP<br />
comprizes a 7S RNA and six polypeptides of 9, 14, 19,<br />
54, 68 and 72 kDa. The SRP54 subunit is a GTPase<br />
to which the signal sequence of a nascent polypeptide<br />
chain binds. The SRP receptor consists of an α and β<br />
subunit both of which are GTPases. The translocation<br />
channel is formed by the Sec61p complex and several<br />
accessory proteins.<br />
Functional analysis of the three translocation-<br />
GTPases, SRP54, SRP receptor α and β<br />
G. Bacher and M. Pool<br />
The GTPase cycle of SRP54 was studied in its functional<br />
context, a ribosome nascent chain complex<br />
(RNC) and membranes containing different components<br />
of the translocation complex (Sec61p, TRAMp,<br />
SRP receptor). It was found that the ribosome stimulates<br />
GTP binding to SRP54 and that the GTP-bound<br />
state of SRP54 is important for high affinity binding<br />
of SRP to the SRP receptor α (Bacher et al. 1996).<br />
Two ribosomal proteins were identified that contact<br />
the GTPase domain and M-domain of SRP54 respectively<br />
(Pool and Dobberstein, in preparation). It is proposed<br />
that one of these proteins allows SRP to scan<br />
the RNC for the presence of a signal sequence and<br />
that the second protein functions in stimulating GDP /<br />
GTP exchange on SRP 54.<br />
The SRP receptor β subunit is anchored in the membrane<br />
and attaches SRP receptor α to the membrane.<br />
We have investigated GTP binding and hydrolysis of<br />
SRP receptor β. We found that SRP receptor β binds<br />
GTP with high affinity and interacts with ribosomes<br />
in the GTP-bound state. Subsequently, the ribosome<br />
increases the GTPase activity of SRP receptor β and<br />
thus functions as a GTPase activating component for<br />
SRP receptor β. We propose that SRP receptor β regulates<br />
the interaction of SRP receptor with the ribosome<br />
and thereby allows SRP receptor α to scan membrane<br />
bound ribosomes for the presence of SRP (Bacher,<br />
Pool and Dobberstein, 1999). Thus the two subunits<br />
of the SRP receptor regulate ribosome binding to the<br />
ER membrane and signal sequence insertion into the<br />
translocon.<br />
61