Microbiology Research - Academic Journals
Microbiology Research - Academic Journals
Microbiology Research - Academic Journals
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
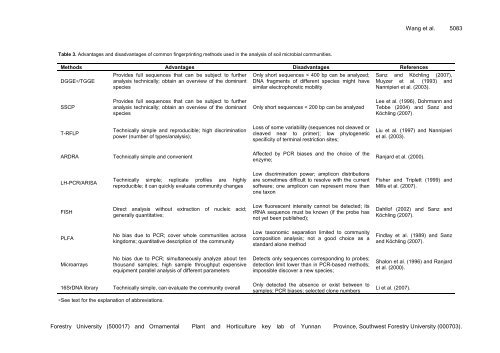
Table 3. Advantages and disadvantages of common fingerprinting methods used in the analysis of soil microbial communities.<br />
Methods Advantages Disadvantages References<br />
DGGE∗/TGGE<br />
SSCP<br />
T-RFLP<br />
Provides full sequences that can be subject to further<br />
analysis technically; obtain an overview of the dominant<br />
species<br />
Provides full sequences that can be subject to further<br />
analysis technically; obtain an overview of the dominant<br />
species<br />
Technically simple and reproducible; high discrimination<br />
power (number of types/analysis);<br />
ARDRA Technically simple and convenient<br />
LH-PCR/ARISA<br />
FISH<br />
PLFA<br />
Microarrays<br />
Technically simple; replicate profiles are highly<br />
reproducible; it can quickly evaluate community changes<br />
Direct analysis without extraction of nucleic acid;<br />
generally quantitative;<br />
No bias due to PCR; cover whole communities across<br />
kingdoms; quantitative description of the community<br />
No bias due to PCR; simultaneously analyze about ten<br />
thousand samples; high sample throughput expensive<br />
equipment parallel analysis of different parameters<br />
16SrDNA library Technically simple, can evaluate the community overall<br />
∗See text for the explanation of abbreviations.<br />
Only short sequences < 400 bp can be analyzed;<br />
DNA fragments of different species might have<br />
similar electrophoretic mobility<br />
Only short sequences < 200 bp can be analyzed<br />
Loss of some variability (sequences not cleaved or<br />
cleaved near to primer); low phylogenetic<br />
specificity of terminal restriction sites;<br />
Affected by PCR biases and the choice of the<br />
enzyme;<br />
Low discrimination power; amplicon distributions<br />
are sometimes difficult to resolve with the current<br />
software; one amplicon can represent more than<br />
one taxon<br />
Low fluorescent intensity cannot be detected; its<br />
rRNA sequence must be known (if the probe has<br />
not yet been published);<br />
Low taxonomic separation limited to community<br />
composition analysis; not a good choice as a<br />
standard alone method<br />
Detects only sequences corresponding to probes;<br />
detection limit lower than in PCR-based methods;<br />
impossible discover a new species;<br />
Only detected the absence or exist between to<br />
samples; PCR biases; selected clone numbers<br />
Wang et al. 5083<br />
Sanz and Köchling (2007),<br />
Muyzer et al. (1993) and<br />
Nannipieri et al. (2003).<br />
Lee et al. (1996), Dohrmann and<br />
Tebbe (2004) and Sanz and<br />
Köchling (2007).<br />
Liu et al. (1997) and Nannipieri<br />
et al. (2003).<br />
Ranjard et al. (2000).<br />
Fisher and Triplett (1999) and<br />
Mills et al. (2007).<br />
Dahllof (2002) and Sanz and<br />
Köchling (2007).<br />
Findlay et al. (1989) and Sanz<br />
and Köchling (2007).<br />
Shalon et al. (1996) and Ranjard<br />
et al. (2000).<br />
Li et al. (2007).<br />
Forestry University (500017) and Ornamental Plant and Horticulture key lab of Yunnan Province, Southwest Forestry University (000703).