Inhibitor SourceBook™ Second Edition
Inhibitor SourceBook™ Second Edition
Inhibitor SourceBook™ Second Edition
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Proteases<br />
Calpain <strong>Inhibitor</strong>s<br />
Calpains belong to a family of calcium-dependent thiolproteases<br />
that proteolyze a wide variety of cytoskeletal,<br />
membrane-associated, and regulatory proteins. Fifteen<br />
gene products of the calpain family are reported in<br />
mammals, which are classified as nine typical and six<br />
atypical calpains. Typical calpains are characterized<br />
by a C-terminal Ca 2+ -binding domain that includes EFhand<br />
motifs, while atypical calpains lack this region,<br />
but often contain additional domains. Calpains do not<br />
generally function as destructive proteases, but act as<br />
calcium-dependent modulators that remove limited<br />
portions of protein substrates. Calpains respond to<br />
Ca 2+ signals by cleaving specific proteins, frequently<br />
components of signaling cascades, thereby irreversibly<br />
modifying their function. Two major isoforms of calpain<br />
are reported in mammals, calpain-1 (m-form) and<br />
calpain-2 (m-form). They are constitutively expressed<br />
in all tissues and differ in their calcium requirement<br />
for activation (~50 mM for calpain-1 and ~500 mM for<br />
calpain-2) and contain several calcium-binding sites,<br />
which allosterically affect the enzyme activity.<br />
Calpain-1 and -2 exhibit about 55-65% sequence<br />
homology and are composed of an 80 kDa and a 30 kDa<br />
subunit. The 80 kDa subunit has the catalytic site and<br />
is unique to each isozyme, whereas the 30 kDa unit is<br />
the regulatory subunit and is common to both calpain-<br />
1 and -2. The 80 kDa subunit consists of four domains<br />
(I-IV) and the 30 kDa unit has 2 domains (V and VI).<br />
Domain I is partially removed during autolysis. Domain<br />
II is the protease domain, which is structurally similar<br />
to the catalytic domain of other cysteine proteases. It<br />
contains a Cys-His-Asn triad characteristic of cysteine<br />
proteases. Domain III exhibits a homology with typical<br />
calmodulin binding proteins and interacts with calcium<br />
binding domains (IV and VI) and frees domain II for<br />
protease activity. Domain IV is a Ca 2+ -binding domain<br />
structurally similar to calmodulin. Domain V contains<br />
a hydrophobic region and is essential for calpain<br />
interaction with membranes. Domain VI has five EFhands.<br />
There are two EF-hand pairs, one pair (EF1-EF2)<br />
displays an ‘open’ conformation and the other (EF3-EF4)<br />
a ‘closed’ conformation. The fifth EF-hand (EF5) is in a<br />
‘closed’ conformation. Both, calpain-1 and -2 associate<br />
non-covalently with domain V and domain VI. Several<br />
putative substrates of calpain-1 and -2 are known<br />
that are cleaved by both isoforms, but with different<br />
efficiencies. Recently, determinants for calpain-1 and<br />
-2 have been analyzed and it is shown that amino acid<br />
preferences extend over 11 residues around the scissile<br />
bond. Calpains prefer Leu, Thr, Val in the P2 position,<br />
Lys, Tyr, Arg in the P1 position, and proline in the region<br />
flanking the P2 - P’1 segment.<br />
Calpain-3, another typical calpain was first described<br />
as a skeletal muscle-specific calpain isoform. However,<br />
subsequent studies have shown its presence in several<br />
other tissues. It is a 94 kDa enzyme that contains<br />
821 amino acids. Structurally calpain-3 is similar<br />
to calpain-1 and -2, however, it has an additional Nterminal<br />
sequence of 20-30 amino acids (NS). Also, its<br />
domain I exhibits less than 20% homology to calpain-1<br />
and -2. Calpain-3 includes two characteristic amino<br />
acid stretches: one between the catalytic cysteine<br />
and histidine (IS1) and the other (IS2) upstream of the<br />
first EF hand motif of calcium-binding domain IV.<br />
A characteristic feature of calpain 3 is that it is not<br />
inhibited by calpastatin. While conventional calpains<br />
localize mainly in the cytosol or at the inner face of<br />
the plasma membrane, calpain-3 is detected within the<br />
nuclei of skeletal muscle cells and in the N2 region of<br />
myofibrils.<br />
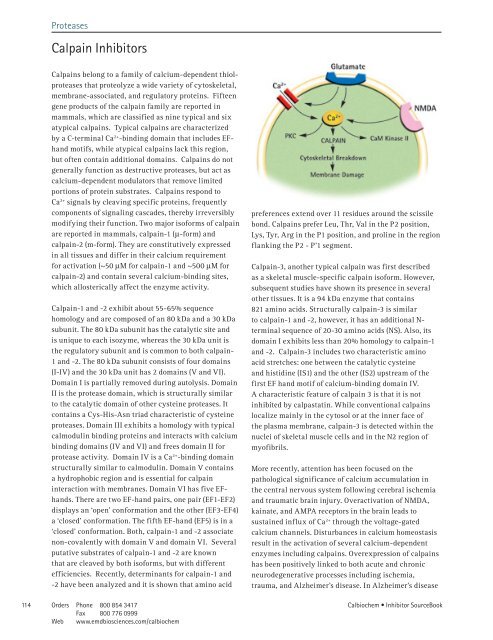
More recently, attention has been focused on the<br />
pathological significance of calcium accumulation in<br />
the central nervous system following cerebral ischemia<br />
and traumatic brain injury. Overactivation of NMDA,<br />
kainate, and AMPA receptors in the brain leads to<br />
sustained influx of Ca 2+ through the voltage-gated<br />
calcium channels. Disturbances in calcium homeostasis<br />
result in the activation of several calcium-dependent<br />
enzymes including calpains. Overexpression of calpains<br />
has been positively linked to both acute and chronic<br />
neurodegenerative processes including ischemia,<br />
trauma, and Alzheimer’s disease. In Alzheimer’s disease<br />
4 Orders Phone 800 854 34 7<br />
Calbiochem • <strong>Inhibitor</strong> SourceBook<br />
Fax 800 776 0999<br />
Web www.emdbiosciences.com/calbiochem