Inhibitor SourceBook™ Second Edition
Inhibitor SourceBook™ Second Edition
Inhibitor SourceBook™ Second Edition
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
Proteases<br />
Proteasome and Ubiquitination <strong>Inhibitor</strong>s<br />
Proteasomes are large multi-subunit complexes,<br />
localized in the nucleus and cytosol that selectively<br />
degrade intracellular proteins. A protein marked for<br />
degradation is covalently attached to multiple molecules<br />
of ubiquitin (Ubq). Four or more Ubq are required to<br />
set a protein for degradation by the proteasome. Ubq<br />
is a highly conserved 76-amino acid (8.6 kDa) protein,<br />
which escorts proteins for rapid hydrolysis to the multicomponent<br />
enzymatic complex, the 26S proteasome. The<br />
proteolytic core of this complex, the 20S proteasome,<br />
contains multiple peptidase activities and functions<br />
as the catalytic machine. This core is composed of 28<br />
subunits arranged in four heptameric, tightly stacked,<br />
rings (a 7 , b 7 , b 7 , a 7 ) to form a cylindrical structure. The<br />
a-subunits make up the two outer and the b-subunits the<br />
two inner rings of the stack. The entrance of substrate<br />
proteins to the active site of the complex is guarded<br />
by the a-subunits that allow access only to unfolded<br />
and extended polypeptides. The proteolytic activity is<br />
confined to the b-subunits. The 19S complex “caps” at<br />
each end of the 20S proteasome help in unfolding protein<br />
substrates. The 19S cap contains subunits with ATPase<br />
activity, subunits that recognize and bind polyubiquitin<br />
chains and putative unfoldases that unfold protein<br />
chains and translocate them into the 20S proteasome.<br />
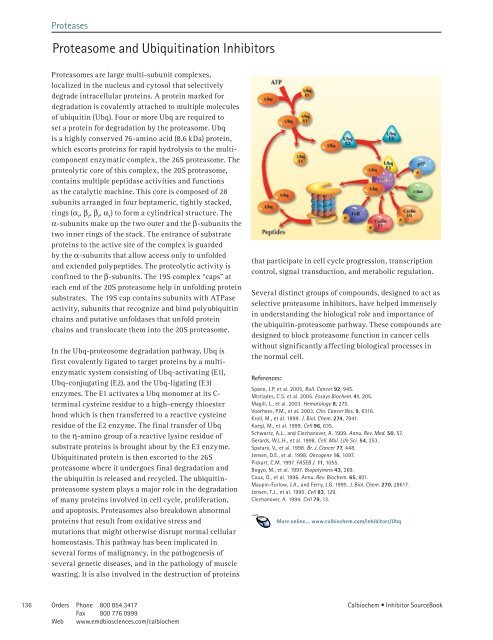
In the Ubq-proteosome degradation pathway, Ubq is<br />
first covalently ligated to target proteins by a multienzymatic<br />
system consisting of Ubq-activating (E1),<br />
Ubq-conjugating (E2), and the Ubq-ligating (E3)<br />
enzymes. The E1 activates a Ubq monomer at its Cterminal<br />
cysteine residue to a high-energy thioester<br />
bond which is then transferred to a reactive cysteine<br />
residue of the E2 enzyme. The final transfer of Ubq<br />
to the h-amino group of a reactive lysine residue of<br />
substrate proteins is brought about by the E3 enzyme.<br />
Ubiquitinated protein is then escorted to the 26S<br />
proteasome where it undergoes final degradation and<br />
the ubiquitin is released and recycled. The ubiquitinproteasome<br />
system plays a major role in the degradation<br />
of many proteins involved in cell cycle, proliferation,<br />
and apoptosis. Proteasomes also breakdown abnormal<br />
proteins that result from oxidative stress and<br />
mutations that might otherwise disrupt normal cellular<br />
homeostasis. This pathway has been implicated in<br />
several forms of malignancy, in the pathogenesis of<br />
several genetic diseases, and in the pathology of muscle<br />
wasting. It is also involved in the destruction of proteins<br />
that participate in cell cycle progression, transcription<br />
control, signal transduction, and metabolic regulation.<br />
Several distinct groups of compounds, designed to act as<br />
selective proteasome inhibitors, have helped immensely<br />
in understanding the biological role and importance of<br />
the ubiquitin-proteasome pathway. These compounds are<br />
designed to block proteasome function in cancer cells<br />
without significantly affecting biological processes in<br />
the normal cell.<br />
References:<br />
Spano, J.P. et al. 2005, Bull. Cancer 92, 945.<br />
Mistiades, C.S. et al. 2005. Essays Biochem. 41, 205.<br />
Magill, L., et al. 2003. Hematology 8, 275.<br />
Voorhees, P.M., et al. 2003. Clin. Cancer Res. 9, 63 6.<br />
Kroll, M., et al. 999. J. Biol. Chem. 274, 794 .<br />
Koegl, M., et al. 999. Cell 96, 635.<br />
Schwartz, A.L., and Ciechanover, A. 999. Annu. Rev. Med. 50, 57.<br />
Gerards, W.L.H., et al. 998. Cell. Mol. Life Sci. 54, 253.<br />
Spataro, V., et al. 998. Br. J. Cancer 77, 448.<br />
Jensen, D.E., et al. 998. Oncogene 16, 097.<br />
Pickart, C.M. 997. FASEB J. 11, 055.<br />
Bogyo, M., et al. 997. Biopolymers 43, 269.<br />
Coux, O., et al. 996. Annu. Rev. Biochem. 65, 80 .<br />
Maupin-Furlow, J.A., and Ferry, J.G. 995. J. Biol. Chem. 270, 286 7.<br />
Jensen, T.J., et al. 995. Cell 83, 29.<br />
Ciechanover, A. 994. Cell 79, 3.<br />
More online... www.calbiochem.com/inhibitors/Ubq<br />
36 Orders Phone 800 854 34 7<br />
Calbiochem • <strong>Inhibitor</strong> SourceBook<br />
Fax 800 776 0999<br />
Web www.emdbiosciences.com/calbiochem