Principles of cell signaling - UT Southwestern
Principles of cell signaling - UT Southwestern
Principles of cell signaling - UT Southwestern
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
39057_ch14_<strong>cell</strong>bio.qxd 8/28/06 5:11 PM Page 622<br />
SUBSTRATE 1<br />
(Protein SER)<br />
H<br />
OH<br />
PRODUCT 1<br />
(Phosphorylated protein)<br />
H<br />
N<br />
C<br />
C<br />
N<br />
C<br />
N<br />
C<br />
C<br />
N<br />
C<br />
H<br />
O<br />
CH 2<br />
H<br />
O<br />
H<br />
O<br />
CH 2<br />
H<br />
O<br />
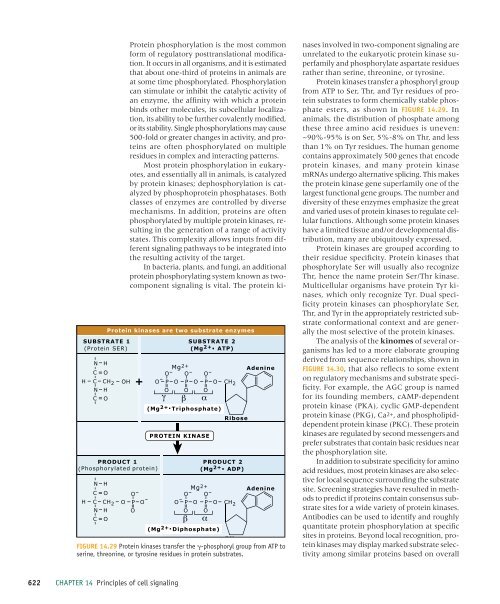
Protein kinases are two substrate enzymes<br />
O - P O -<br />
O<br />
+<br />
O - P O<br />
O<br />
SUBSTRATE<br />
(Mg 2+. 2<br />
ATP)<br />
Mg 2+<br />
O - P O<br />
O<br />
O - O -<br />
O - P O<br />
O<br />
O - P O<br />
O<br />
CH 2<br />
(Mg 2+. Triphosphate)<br />
Ribose<br />
PROTEIN KINASE<br />
PRODUCT<br />
(Mg 2+. 2<br />
ADP)<br />
Mg 2+<br />
O - P O<br />
O<br />
CH 2<br />
Adenine<br />
Adenine<br />
O<br />
(Mg 2+. Diphosphate)<br />
Rib<br />
FIGURE 14.29 Protein kinases transfer the -phosphoryl group from ATP to<br />
serine, threonine, or tyrosine residues in protein substrates.<br />
Protein phosphorylation is the most common<br />
form <strong>of</strong> regulatory posttranslational modification.<br />
It occurs in all organisms, and it is estimated<br />
that about one-third <strong>of</strong> proteins in animals are<br />
at some time phosphorylated. Phosphorylation<br />
can stimulate or inhibit the catalytic activity <strong>of</strong><br />
an enzyme, the affinity with which a protein<br />
binds other molecules, its sub<strong>cell</strong>ular localization,<br />
its ability to be further covalently modified,<br />
or its stability. Single phosphorylations may cause<br />
500-fold or greater changes in activity, and proteins<br />
are <strong>of</strong>ten phosphorylated on multiple<br />
residues in complex and interacting patterns.<br />
Most protein phosphorylation in eukaryotes,<br />
and essentially all in animals, is catalyzed<br />
by protein kinases; dephosphorylation is catalyzed<br />
by phosphoprotein phosphatases. Both<br />
classes <strong>of</strong> enzymes are controlled by diverse<br />
mechanisms. In addition, proteins are <strong>of</strong>ten<br />
phosphorylated by multiple protein kinases, resulting<br />
in the generation <strong>of</strong> a range <strong>of</strong> activity<br />
states. This complexity allows inputs from different<br />
<strong>signaling</strong> pathways to be integrated into<br />
the resulting activity <strong>of</strong> the target.<br />
In bacteria, plants, and fungi, an additional<br />
protein phosphorylating system known as twocomponent<br />
<strong>signaling</strong> is vital. The protein kinases<br />
involved in two-component <strong>signaling</strong> are<br />
unrelated to the eukaryotic protein kinase superfamily<br />
and phosphorylate aspartate residues<br />
rather than serine, threonine, or tyrosine.<br />
Protein kinases transfer a phosphoryl group<br />
from ATP to Ser, Thr, and Tyr residues <strong>of</strong> protein<br />
substrates to form chemically stable phosphate<br />
esters, as shown in FIGURE 14.29. In<br />
animals, the distribution <strong>of</strong> phosphate among<br />
these three amino acid residues is uneven:<br />
~90%-95% is on Ser, 5%-8% on Thr, and less<br />
than 1% on Tyr residues. The human genome<br />
contains approximately 500 genes that encode<br />
protein kinases, and many protein kinase<br />
mRNAs undergo alternative splicing. This makes<br />
the protein kinase gene superfamily one <strong>of</strong> the<br />
largest functional gene groups. The number and<br />
diversity <strong>of</strong> these enzymes emphasize the great<br />
and varied uses <strong>of</strong> protein kinases to regulate <strong>cell</strong>ular<br />
functions. Although some protein kinases<br />
have a limited tissue and/or developmental distribution,<br />
many are ubiquitously expressed.<br />
Protein kinases are grouped according to<br />
their residue specificity. Protein kinases that<br />
phosphorylate Ser will usually also recognize<br />
Thr, hence the name protein Ser/Thr kinase.<br />
Multi<strong>cell</strong>ular organisms have protein Tyr kinases,<br />
which only recognize Tyr. Dual specificity<br />
protein kinases can phosphorylate Ser,<br />
Thr, and Tyr in the appropriately restricted substrate<br />
conformational context and are generally<br />
the most selective <strong>of</strong> the protein kinases.<br />
The analysis <strong>of</strong> the kinomes <strong>of</strong> several organisms<br />
has led to a more elaborate grouping<br />
derived from sequence relationships, shown in<br />
FIGURE 14.30, that also reflects to some extent<br />
on regulatory mechanisms and substrate specificity.<br />
For example, the AGC group is named<br />
for its founding members, cAMP-dependent<br />
protein kinase (PKA), cyclic GMP-dependent<br />
protein kinase (PKG), Ca2+, and phospholipiddependent<br />
protein kinase (PKC). These protein<br />
kinases are regulated by second messengers and<br />
prefer substrates that contain basic residues near<br />
the phosphorylation site.<br />
In addition to substrate specificity for amino<br />
acid residues, most protein kinases are also selective<br />
for local sequence surrounding the substrate<br />
site. Screening strategies have resulted in methods<br />
to predict if proteins contain consensus substrate<br />
sites for a wide variety <strong>of</strong> protein kinases.<br />
Antibodies can be used to identify and roughly<br />
quantitate protein phosphorylation at specific<br />
sites in proteins. Beyond local recognition, protein<br />
kinases may display marked substrate selectivity<br />
among similar proteins based on overall<br />
622 CHAPTER 14 <strong>Principles</strong> <strong>of</strong> <strong>cell</strong> <strong>signaling</strong>