Principles of cell signaling - UT Southwestern
Principles of cell signaling - UT Southwestern
Principles of cell signaling - UT Southwestern
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
39057_ch14_<strong>cell</strong>bio.qxd 8/28/06 5:11 PM Page 631<br />
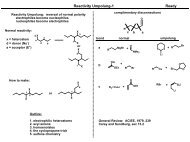
Structure and regulation <strong>of</strong> Src<br />
INACTIVE<br />
ACTIVE<br />
KINASE<br />
DOMAIN<br />
SH3<br />
SH2<br />
P<br />
P<br />
Tyr527<br />
FIGURE 14.37 The structures <strong>of</strong> inactive and active Src are compared. The inactive<br />
protein is autoinhibited by binding to its own SH2 and SH3 domains.<br />
The SH2 domain binds to phosphorylated Tyr527. The SH3 domain binds to a<br />
noncanonical SH3-binding motif on the opposite side <strong>of</strong> the kinase domain<br />
active site. In contrast to the steric inhibition <strong>of</strong> PKA caused by its R subunit,<br />
inhibition <strong>of</strong> Src by its SH2 and SH3 domains is allosteric. In the active structure<br />
the SH2 and SH3 domains are not bound to the kinase domain and are<br />
available for heterologous interactions. Structures generated from Protein Data<br />
Base files 1FMK and 1Y57.<br />
14.32<br />
MAPKs are central to<br />
many <strong>signaling</strong> pathways<br />
Key concepts<br />
• MAPKs are activated by Tyr and Thr<br />
phosphorylation.<br />
• The requirement for two phosphorylations creates<br />
a <strong>signaling</strong> threshold.<br />
• The ERK1/2 MAPK pathway is usually regulated<br />
through Ras.<br />
Mitogen-activated protein kinases (MAPKs) are<br />
present in all eukaryotes. They are among the<br />
most common multifunctional protein kinases<br />
mediating <strong>cell</strong>ular regulatory events in response<br />
to many ligands and other stimuli. MAPKs are<br />
activated by protein kinase cascades consisting<br />
<strong>of</strong> at least three protein kinases acting sequentially,<br />
as illustrated in FIGURE 14.38. Activation<br />
<strong>of</strong> a MAPK is catalyzed by a MAPK kinase<br />
(MAP2K), which is itself activated by phosphorylation<br />
by a MAPK kinase kinase (MAP3K).<br />
MAP3Ks are activated by a variety <strong>of</strong> mechanisms<br />
including phosphorylation by MAP4Ks,<br />
oligomerization, and binding to activators such<br />
as small G proteins.<br />
MAP2Ks are activated by phosphorylation<br />
on two Ser/Thr residues; MAP2Ks then activate<br />
MAPKs by dual phosphorylation on Tyr<br />
and Thr residues (Figure 14.30). Each MAP2K<br />
phosphorylates a limited set <strong>of</strong> MAPKs and few<br />
or no other substrates. The great specificity <strong>of</strong><br />
MAP2Ks is one means <strong>of</strong> insulating MAPKs<br />
from activation by inappropriate signals. Both<br />
Tyr and Thr phosphorylations are required for<br />
maximum MAPK enzymatic activity.<br />
Studies on the MAPK ERK2 led to an understanding<br />
<strong>of</strong> the events induced by phosphorylation<br />
that are important for increased activity.<br />
Conformational changes include refolding <strong>of</strong><br />
the activation loop to improve substrate positioning<br />
and realignment <strong>of</strong> catalytic residues;<br />
this is most obvious in the repositioning <strong>of</strong> <br />
helix C, which contains a Glu involved in phosphoryl<br />
transfer.<br />
Amplification occurs moving down the cascade<br />
from the MAP3K to the MAP2K step because<br />
the MAP2Ks are much more abundant<br />
than the MAP3Ks. The MAP2K to MAPK step<br />
may also amplify the signal if the MAPK is present<br />
in excess <strong>of</strong> the MAP2K. In addition, the<br />
phosphorylation <strong>of</strong> a MAPK by a MAP2K on a<br />
14.32 MAPKs are central to many <strong>signaling</strong> pathways 631