Principles of cell signaling - UT Southwestern
Principles of cell signaling - UT Southwestern
Principles of cell signaling - UT Southwestern
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
39057_ch14_<strong>cell</strong>bio.qxd 8/28/06 5:11 PM Page 596<br />
RECEPTORS<br />
TRANSDUCERS<br />
EFFECTORS<br />
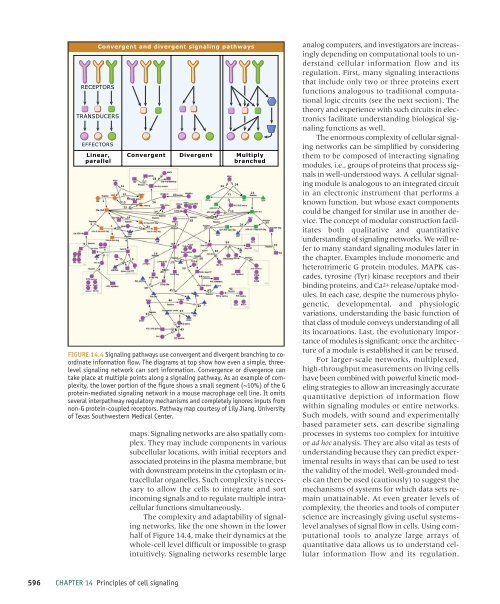
Convergent and divergent <strong>signaling</strong> pathways<br />
Linear,<br />
parallel<br />
Convergent Divergent Multiply<br />
branched<br />
FIGURE 14.4 Signaling pathways use convergent and divergent branching to coordinate<br />
information flow. The diagrams at top show how even a simple, threelevel<br />
<strong>signaling</strong> network can sort information. Convergence or divergence can<br />
take place at multiple points along a <strong>signaling</strong> pathway. As an example <strong>of</strong> complexity,<br />
the lower portion <strong>of</strong> the figure shows a small segment (~10%) <strong>of</strong> the G<br />
protein-mediated <strong>signaling</strong> network in a mouse macrophage <strong>cell</strong> line. It omits<br />
several interpathway regulatory mechanisms and completely ignores inputs from<br />
non-G protein-coupled receptors. Pathway map courtesy <strong>of</strong> Lily Jiang, University<br />
<strong>of</strong> Texas <strong>Southwestern</strong> Medical Center.<br />
maps. Signaling networks are also spatially complex.<br />
They may include components in various<br />
sub<strong>cell</strong>ular locations, with initial receptors and<br />
associated proteins in the plasma membrane, but<br />
with downstream proteins in the cytoplasm or intra<strong>cell</strong>ular<br />
organelles. Such complexity is necessary<br />
to allow the <strong>cell</strong>s to integrate and sort<br />
incoming signals and to regulate multiple intra<strong>cell</strong>ular<br />
functions simultaneously.<br />
The complexity and adaptability <strong>of</strong> <strong>signaling</strong><br />
networks, like the one shown in the lower<br />
half <strong>of</strong> Figure 14.4, make their dynamics at the<br />
whole-<strong>cell</strong> level difficult or impossible to grasp<br />
intuitively. Signaling networks resemble large<br />
analog computers, and investigators are increasingly<br />
depending on computational tools to understand<br />
<strong>cell</strong>ular information flow and its<br />
regulation. First, many <strong>signaling</strong> interactions<br />
that include only two or three proteins exert<br />
functions analogous to traditional computational<br />
logic circuits (see the next section). The<br />
theory and experience with such circuits in electronics<br />
facilitate understanding biological <strong>signaling</strong><br />
functions as well.<br />
The enormous complexity <strong>of</strong> <strong>cell</strong>ular <strong>signaling</strong><br />
networks can be simplified by considering<br />
them to be composed <strong>of</strong> interacting <strong>signaling</strong><br />
modules, i.e., groups <strong>of</strong> proteins that process signals<br />
in well-understood ways. A <strong>cell</strong>ular <strong>signaling</strong><br />
module is analogous to an integrated circuit<br />
in an electronic instrument that performs a<br />
known function, but whose exact components<br />
could be changed for similar use in another device.<br />
The concept <strong>of</strong> modular construction facilitates<br />
both qualitative and quantitative<br />
understanding <strong>of</strong> <strong>signaling</strong> networks. We will refer<br />
to many standard <strong>signaling</strong> modules later in<br />
the chapter. Examples include monomeric and<br />
heterotrimeric G protein modules, MAPK cascades,<br />
tyrosine (Tyr) kinase receptors and their<br />
binding proteins, and Ca2+ release/uptake modules.<br />
In each case, despite the numerous phylogenetic,<br />
developmental, and physiologic<br />
variations, understanding the basic function <strong>of</strong><br />
that class <strong>of</strong> module conveys understanding <strong>of</strong> all<br />
its incarnations. Last, the evolutionary importance<br />
<strong>of</strong> modules is significant; once the architecture<br />
<strong>of</strong> a module is established it can be reused.<br />
For larger-scale networks, multiplexed,<br />
high-throughput measurements on living <strong>cell</strong>s<br />
have been combined with powerful kinetic modeling<br />
strategies to allow an increasingly accurate<br />
quantitative depiction <strong>of</strong> information flow<br />
within <strong>signaling</strong> modules or entire networks.<br />
Such models, with sound and experimentally<br />
based parameter sets, can describe <strong>signaling</strong><br />
processes in systems too complex for intuitive<br />
or ad hoc analysis. They are also vital as tests <strong>of</strong><br />
understanding because they can predict experimental<br />
results in ways that can be used to test<br />
the validity <strong>of</strong> the model. Well-grounded models<br />
can then be used (cautiously) to suggest the<br />
mechanisms <strong>of</strong> systems for which data sets remain<br />
unattainable. At even greater levels <strong>of</strong><br />
complexity, the theories and tools <strong>of</strong> computer<br />
science are increasingly giving useful systemslevel<br />
analyses <strong>of</strong> signal flow in <strong>cell</strong>s. Using computational<br />
tools to analyze large arrays <strong>of</strong><br />
quantitative data allows us to understand <strong>cell</strong>ular<br />
information flow and its regulation.<br />
596 CHAPTER 14 <strong>Principles</strong> <strong>of</strong> <strong>cell</strong> <strong>signaling</strong>