- Page 2 and 3:
Peptides 2010Tales of PeptidesProce
- Page 4 and 5:
Peptides 2010Tales of PeptidesProce
- Page 8 and 9:
IntroductionWe had the distinct hon
- Page 10:
Scientific programIn planning the 3
- Page 13 and 14:
31 st EUROPEAN PEPTIDE SYMPOSIUMSep
- Page 19 and 20:
published by John Wiley & Sons as a
- Page 21 and 22:
The Josef Rudinger AwardThis award
- Page 23 and 24:
Young Investigators' SymposiumThe y
- Page 25 and 26:
xxiv
- Page 27 and 28:
Design of Peptidyl-Inhibitors for G
- Page 29 and 30:
Synthetic Antifreeze Glycopeptide A
- Page 31 and 32:
Synthesis and Oxidative Folding of
- Page 33 and 34:
Convergent Syntheses of Huprp106-12
- Page 35 and 36:
Bactericidal Activity of Small Beta
- Page 37 and 38:
Bradykinin Analogues Acylated on Th
- Page 39 and 40:
Antimicrobial Activity of Small 3-(
- Page 41 and 42:
Interaction of Curcumin with A-Synu
- Page 43 and 44:
Fluorescent and Luminescent Fusion
- Page 45 and 46:
Cellular Expression of the Human An
- Page 47:
2D IR Spectroscopy of Oligopeptides
- Page 50 and 51:
large, non-redundant subset of prot
- Page 52 and 53:
The aberrantly glucosylated peptide
- Page 54 and 55:
Table 1. Proline cis/trans ratios o
- Page 56 and 57:
Table 1. Yields, UV-vis absorption
- Page 58 and 59:
Table 1. Purity of the targeted tri
- Page 60 and 61:
processes are blocked. Interestingl
- Page 62 and 63:
(tb4 n ,1 m )MTII(tb1 n ,4 m )MTIIF
- Page 64 and 65:
Fig. 2. a) Part of the 400 MHz HR-M
- Page 66 and 67:
Once printed, the amino acid partic
- Page 68 and 69:
A) PSA, few seconds73B) PSA, 45 min
- Page 70 and 71:
short β strands. In contrast, nume
- Page 72 and 73:
neg. control Cs9-Rhd MM-218Fig. 2.
- Page 74 and 75:
Table 1. The general structure of t
- Page 76 and 77:
% ID in the liver 1 h p.i.100908070
- Page 78 and 79:
Peptidyl phosphoranes were first re
- Page 80 and 81:
obtained from the same sardines and
- Page 82 and 83:
MccJ25 variants were produced by si
- Page 84 and 85:
Fig. 2. Proposed signaling mechanis
- Page 86 and 87:
Table 1. mCD4-HS12 antiviral activi
- Page 89 and 90:
Proceedings of the 31 st European P
- Page 91 and 92:
Proceedings of the 31 st European P
- Page 93 and 94:
Proceedings of the 31 st European P
- Page 95 and 96:
Proceedings of the 31 st European P
- Page 97 and 98:
Proceedings of the 31 st European P
- Page 99 and 100:
Proceedings of the 31 st European P
- Page 101 and 102:
Proceedings of the 31 st European P
- Page 103 and 104:
Proceedings of the 31 st European P
- Page 105 and 106:
Proceedings of the 31 st European P
- Page 107 and 108:
Proceedings of the 31 st European P
- Page 109 and 110:
RT: 0,00 - 5,9936000034000032000030
- Page 111 and 112:
Proceedings of the 31 st European P
- Page 113 and 114:
Proceedings of the 31 st European P
- Page 115 and 116:
Proceedings of the 31 st European P
- Page 117 and 118:
Proceedings of the 31 st European P
- Page 119 and 120:
Proceedings of the 31 st European P
- Page 121 and 122:
Proceedings of the 31 st European P
- Page 123 and 124:
Proceedings of the 31 st European P
- Page 125 and 126:
Proceedings of the 31 st European P
- Page 127 and 128:
Proceedings of the 31 st European P
- Page 129 and 130:
Proceedings of the 31 st European P
- Page 131 and 132:
Proceedings of the 31 st European P
- Page 133 and 134:
Proceedings of the 31 st European P
- Page 135 and 136:
Proceedings of the 31 st European P
- Page 137 and 138:
Proceedings of the 31 st European P
- Page 139 and 140:
Proceedings of the 31 st European P
- Page 141 and 142:
Proceedings of the 31 st European P
- Page 143 and 144:
Proceedings of the 31 st European P
- Page 145 and 146:
Proceedings of the 31 st European P
- Page 147 and 148:
Proceedings of the 31 st European P
- Page 149 and 150:
Proceedings of the 31 st European P
- Page 151 and 152:
Proceedings of the 31 st European P
- Page 153 and 154:
Proceedings of the 31 st European P
- Page 155 and 156:
Proceedings of the 31 st European P
- Page 157 and 158:
Proceedings of the 31 st European P
- Page 159 and 160:
Proceedings of the 31 st European P
- Page 161 and 162:
Proceedings of the 31 st European P
- Page 163 and 164:
Proceedings of the 31 st European P
- Page 165 and 166:
Proceedings of the 31 st European P
- Page 167 and 168:
Proceedings of the 31 st European P
- Page 169 and 170:
Proceedings of the 31 st European P
- Page 171 and 172:
Proceedings of the 31 st European P
- Page 173 and 174:
Proceedings of the 31 st European P
- Page 175 and 176:
Proceedings of the 31 st European P
- Page 177 and 178:
Proceedings of the 31 st European P
- Page 179 and 180:
Proceedings of the 31 st European P
- Page 181 and 182:
Proceedings of the 31 st European P
- Page 183 and 184:
Proceedings of the 31 st European P
- Page 185 and 186:
Proceedings of the 31 st European P
- Page 187 and 188:
Proceedings of the 31 st European P
- Page 189 and 190:
Proceedings of the 31 st European P
- Page 191 and 192:
Proceedings of the 31 st European P
- Page 193 and 194:
Proceedings of the 31 st European P
- Page 195 and 196:
Proceedings of the 31 st European P
- Page 197 and 198:
Proceedings of the 31 st European P
- Page 199 and 200:
Proceedings of the 31 st European P
- Page 201 and 202:
Proceedings of the 31 st European P
- Page 203 and 204:
Proceedings of the 31 st European P
- Page 205 and 206:
Proceedings of the 31 st European P
- Page 207 and 208:
Proceedings of the 31 st European P
- Page 209 and 210:
Proceedings of the 31 st European P
- Page 211 and 212:
Proceedings of the 31 st European P
- Page 213 and 214:
Proceedings of the 31 st European P
- Page 215 and 216:
Proceedings of the 31 st European P
- Page 217 and 218:
Proceedings of the 31 st European P
- Page 219 and 220:
Proceedings of the 31 st European P
- Page 221 and 222:
Proceedings of the 31 st European P
- Page 223 and 224:
Proceedings of the 31 st European P
- Page 225 and 226:
Proceedings of the 31 st European P
- Page 227 and 228:
Proceedings of the 31 st European P
- Page 229 and 230:
Proceedings of the 31 st European P
- Page 231 and 232:
Proceedings of the 31 st European P
- Page 233 and 234:
Proceedings of the 31 st European P
- Page 235 and 236:
Proceedings of the 31 st European P
- Page 237 and 238:
Proceedings of the 31 st European P
- Page 239 and 240:
Proceedings of the 31 st European P
- Page 241 and 242:
Proceedings of the 31 st European P
- Page 243 and 244:
Proceedings of the 31 st European P
- Page 245 and 246:
Proceedings of the 31 st European P
- Page 247 and 248:
Proceedings of the 31 st European P
- Page 249 and 250:
Proceedings of the 31 st European P
- Page 251 and 252:
Proceedings of the 31 st European P
- Page 253 and 254:
Proceedings of the 31 st European P
- Page 255 and 256:
Proceedings of the 31 st European P
- Page 257 and 258:
Proceedings of the 31 st European P
- Page 259 and 260:
Proceedings of the 31 st European P
- Page 261 and 262:
Proceedings of the 31 st European P
- Page 263 and 264:
Proceedings of the 31 st European P
- Page 265 and 266:
Proceedings of the 31 st European P
- Page 267 and 268:
Proceedings of the 31 st European P
- Page 269 and 270:
Proceedings of the 31 st European P
- Page 271 and 272:
Proceedings of the 31 st European P
- Page 273 and 274:
Proceedings of the 31 st European P
- Page 275 and 276:
Proceedings of the 31 st European P
- Page 277 and 278:
Proceedings of the 31 st European P
- Page 279 and 280:
Proceedings of the 31 st European P
- Page 281 and 282:
Proceedings of the 31 st European P
- Page 283 and 284:
Proceedings of the 31 st European P
- Page 285 and 286:
Proceedings of the 31 st European P
- Page 287 and 288:
Proceedings of the 31 st European P
- Page 289 and 290:
Proceedings of the 31 st European P
- Page 291 and 292: Proceedings of the 31 st European P
- Page 293 and 294: Proceedings of the 31 st European P
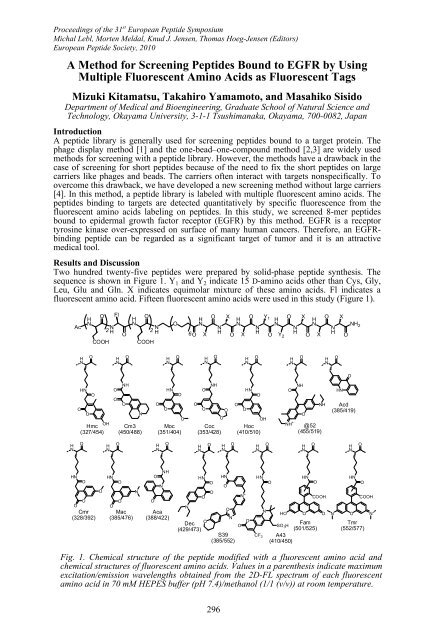
- Page 295 and 296: Proceedings of the 31 st European P
- Page 297 and 298: Proceedings of the 31 st European P
- Page 299 and 300: Proceedings of the 31 st European P
- Page 301 and 302: Proceedings of the 31 st European P
- Page 303 and 304: Proceedings of the 31 st European P
- Page 305 and 306: Proceedings of the 31 st European P
- Page 307 and 308: Proceedings of the 31 st European P
- Page 309 and 310: Proceedings of the 31 st European P
- Page 311 and 312: Proceedings of the 31 st European P
- Page 313 and 314: Proceedings of the 31 st European P
- Page 315 and 316: Proceedings of the 31 st European P
- Page 317 and 318: Proceedings of the 31 st European P
- Page 319 and 320: Proceedings of the 31 st European P
- Page 321 and 322: Proceedings of the 31 st European P
- Page 323 and 324: Proceedings of the 31 st European P
- Page 325 and 326: Proceedings of the 31 st European P
- Page 327 and 328: Proceedings of the 31 st European P
- Page 329 and 330: Proceedings of the 31 st European P
- Page 331 and 332: Proceedings of the 31 st European P
- Page 333 and 334: Proceedings of the 31 st European P
- Page 335 and 336: Proceedings of the 31 st European P
- Page 337 and 338: Proceedings of the 31 st European P
- Page 339 and 340: Proceedings of the 31 st European P
- Page 341: Proceedings of the 31 st European P
- Page 345 and 346: Proceedings of the 31 st European P
- Page 347 and 348: Proceedings of the 31 st European P
- Page 349 and 350: Proceedings of the 31 st European P
- Page 351 and 352: Proceedings of the 31 st European P
- Page 353 and 354: Proceedings of the 31 st European P
- Page 355 and 356: Proceedings of the 31 st European P
- Page 357 and 358: Proceedings of the 31 st European P
- Page 359 and 360: Proceedings of the 31 st European P
- Page 361 and 362: Proceedings of the 31 st European P
- Page 363 and 364: Proceedings of the 31 st European P
- Page 365 and 366: Proceedings of the 31 st European P
- Page 367 and 368: Proceedings of the 31 st European P
- Page 369 and 370: Proceedings of the 31 st European P
- Page 371 and 372: Proceedings of the 31 st European P
- Page 373 and 374: Proceedings of the 31 st European P
- Page 375 and 376: Proceedings of the 31 st European P
- Page 377 and 378: Proceedings of the 31 st European P
- Page 379 and 380: Proceedings of the 31 st European P
- Page 381 and 382: Proceedings of the 31 st European P
- Page 383 and 384: Proceedings of the 31 st European P
- Page 385 and 386: Proceedings of the 31 st European P
- Page 387 and 388: Proceedings of the 31 st European P
- Page 389 and 390: Proceedings of the 31 st European P
- Page 391 and 392: Proceedings of the 31 st European P
- Page 393 and 394:
Proceedings of the 31 st European P
- Page 395 and 396:
Proceedings of the 31 st European P
- Page 397 and 398:
Proceedings of the 31 st European P
- Page 399 and 400:
Proceedings of the 31 st European P
- Page 401 and 402:
Proceedings of the 31 st European P
- Page 403 and 404:
Proceedings of the 31 st European P
- Page 405 and 406:
Proceedings of the 31 st European P
- Page 407 and 408:
Proceedings of the 31 st European P
- Page 409 and 410:
Proceedings of the 31 st European P
- Page 411 and 412:
Proceedings of the 31 st European P
- Page 413 and 414:
Proceedings of the 31 st European P
- Page 415 and 416:
Proceedings of the 31 st European P
- Page 417 and 418:
Proceedings of the 31 st European P
- Page 419 and 420:
Proceedings of the 31 st European P
- Page 421 and 422:
Proceedings of the 31 st European P
- Page 423 and 424:
Proceedings of the 31 st European P
- Page 425 and 426:
Proceedings of the 31 st European P
- Page 427 and 428:
Proceedings of the 31 st European P
- Page 429 and 430:
Proceedings of the 31 st European P
- Page 431 and 432:
Proceedings of the 31 st European P
- Page 433 and 434:
Proceedings of the 31 st European P
- Page 435 and 436:
Proceedings of the 31 st European P
- Page 437 and 438:
Proceedings of the 31 st European P
- Page 439 and 440:
Proceedings of the 31 st European P
- Page 441 and 442:
Proceedings of the 31 st European P
- Page 443 and 444:
Proceedings of the 31 st European P
- Page 445 and 446:
Proceedings of the 31 st European P
- Page 447 and 448:
Proceedings of the 31 st European P
- Page 449 and 450:
Proceedings of the 31 st European P
- Page 451 and 452:
Proceedings of the 31 st European P
- Page 453 and 454:
Proceedings of the 31 st European P
- Page 455 and 456:
Proceedings of the 31 st European P
- Page 457 and 458:
Proceedings of the 31 st European P
- Page 459 and 460:
Proceedings of the 31 st European P
- Page 461 and 462:
Proceedings of the 31 st European P
- Page 463 and 464:
Proceedings of the 31 st European P
- Page 465 and 466:
Proceedings of the 31 st European P
- Page 467 and 468:
Proceedings of the 31 st European P
- Page 469 and 470:
Proceedings of the 31 st European P
- Page 471 and 472:
Proceedings of the 31 st European P
- Page 473 and 474:
Proceedings of the 31 st European P
- Page 475 and 476:
Proceedings of the 31 st European P
- Page 477 and 478:
Proceedings of the 31 st European P
- Page 479 and 480:
Proceedings of the 31 st European P
- Page 481 and 482:
Proceedings of the 31 st European P
- Page 483 and 484:
Proceedings of the 31 st European P
- Page 485 and 486:
Proceedings of the 31 st European P
- Page 487 and 488:
Proceedings of the 31 st European P
- Page 489 and 490:
Proceedings of the 31 st European P
- Page 491 and 492:
Proceedings of the 31 st European P
- Page 493 and 494:
Proceedings of the 31 st European P
- Page 495 and 496:
Proceedings of the 31 st European P
- Page 497 and 498:
Proceedings of the 31 st European P
- Page 499 and 500:
Proceedings of the 31 st European P
- Page 501 and 502:
Proceedings of the 31 st European P
- Page 503 and 504:
Proceedings of the 31 st European P
- Page 505 and 506:
Proceedings of the 31 st European P
- Page 507 and 508:
Proceedings of the 31 st European P
- Page 509 and 510:
Proceedings of the 31 st European P
- Page 511 and 512:
Proceedings of the 31 st European P
- Page 513 and 514:
Proceedings of the 31 st European P
- Page 515 and 516:
Proceedings of the 31 st European P
- Page 517 and 518:
Proceedings of the 31 st European P
- Page 519 and 520:
Proceedings of the 31 st European P
- Page 521 and 522:
Proceedings of the 31 st European P
- Page 523 and 524:
Proceedings of the 31 st European P
- Page 525 and 526:
Proceedings of the 31 st European P
- Page 527 and 528:
Proceedings of the 31 st European P
- Page 529 and 530:
Proceedings of the 31 st European P
- Page 531 and 532:
Proceedings of the 31 st European P
- Page 533 and 534:
Proceedings of the 31 st European P
- Page 535 and 536:
Proceedings of the 31 st European P
- Page 537 and 538:
Proceedings of the 31 st European P
- Page 539 and 540:
Proceedings of the 31 st European P
- Page 541 and 542:
Proceedings of the 31 st European P
- Page 543 and 544:
Proceedings of the 31 st European P
- Page 545 and 546:
Proceedings of the 31 st European P
- Page 547 and 548:
Proceedings of the 31 st European P
- Page 549 and 550:
Proceedings of the 31 st European P
- Page 551 and 552:
Proceedings of the 31 st European P
- Page 553 and 554:
Proceedings of the 31 st European P
- Page 555 and 556:
Proceedings of the 31 st European P
- Page 557 and 558:
Proceedings of the 31 st European P
- Page 559 and 560:
Proceedings of the 31 st European P
- Page 561 and 562:
Proceedings of the 31 st European P
- Page 563 and 564:
Proceedings of the 31 st European P
- Page 565 and 566:
Proceedings of the 31 st European P
- Page 567 and 568:
Proceedings of the 31 st European P
- Page 569 and 570:
Proceedings of the 31 st European P
- Page 571 and 572:
Proceedings of the 31 st European P
- Page 573 and 574:
Proceedings of the 31 st European P
- Page 575 and 576:
Proceedings of the 31 st European P
- Page 577 and 578:
Proceedings of the 31 st European P
- Page 579 and 580:
Proceedings of the 31 st European P
- Page 581 and 582:
Proceedings of the 31 st European P
- Page 583 and 584:
Proceedings of the 31 st European P
- Page 585 and 586:
Proceedings of the 31 st European P
- Page 587 and 588:
Proceedings of the 31 st European P
- Page 589 and 590:
Proceedings of the 31 st European P
- Page 591 and 592:
Proceedings of the 31 st European P
- Page 593 and 594:
Proceedings of the 31 st European P
- Page 595 and 596:
Proceedings of the 31 st European P
- Page 597 and 598:
Proceedings of the 31 st European P
- Page 599 and 600:
Proceedings of the 31 st European P
- Page 601 and 602:
Proceedings of the 31 st European P
- Page 603 and 604:
Proceedings of the 31 st European P
- Page 605 and 606:
Proceedings of the 31 st European P
- Page 607 and 608:
Proceedings of the 31 st European P
- Page 609 and 610:
Proceedings of the 31 st European P
- Page 611 and 612:
Proceedings of the 31 st European P
- Page 613 and 614:
Proceedings of the 31 st European P
- Page 615 and 616:
Proceedings of the 31 st European P
- Page 617 and 618:
Proceedings of the 31 st European P
- Page 619 and 620:
Proceedings of the 31 st European P
- Page 621 and 622:
Proceedings of the 31 st European P
- Page 623 and 624:
Proceedings of the 31 st European P
- Page 625 and 626:
Proceedings of the 31 st European P
- Page 627 and 628:
Proceedings of the 31 st European P
- Page 629 and 630:
Proceedings of the 31 st European P
- Page 631 and 632:
Proceedings of the 31 st European P
- Page 633 and 634:
Proceedings of the 31 st European P
- Page 635 and 636:
Proceedings of the 31 st European P
- Page 637 and 638:
Proceedings of the 31 st European P
- Page 639 and 640:
Proceedings of the 31 st European P
- Page 641 and 642:
Proceedings of the 31 st European P
- Page 643 and 644:
Proceedings of the 31 st European P
- Page 645 and 646:
Proceedings of the 31 st European P
- Page 647 and 648:
Proceedings of the 31 st European P
- Page 649 and 650:
Proceedings of the 31 st European P
- Page 651 and 652:
Proceedings of the 31 st European P
- Page 653 and 654:
Proceedings of the 31 st European P
- Page 655 and 656:
Proceedings of the 31 st European P
- Page 657 and 658:
Proceedings of the 31 st European P
- Page 659 and 660:
Proceedings of the 31 st European P
- Page 661 and 662:
Proceedings of the 31 st European P
- Page 663 and 664:
Proceedings of the 31 st European P
- Page 665 and 666:
Proceedings of the 31 st European P
- Page 667 and 668:
Proceedings of the 31 st European P
- Page 669 and 670:
Proceedings of the 31 st European P
- Page 671 and 672:
Proceedings of the 31 st European P
- Page 673 and 674:
Proceedings of the 31 st European P
- Page 675 and 676:
Proceedings of the 31 st European P
- Page 677 and 678:
Proceedings of the 31 st European P
- Page 679 and 680:
Subject index(S)-2-(1-adamantyl)gly
- Page 681 and 682:
chip 18chiral triazine condensing r
- Page 683 and 684:
heat stress 348helical conformation
- Page 685 and 686:
O-acyl isopeptide method 112obesity
- Page 687 and 688:
SH2-ligands 210short collagen-relat
- Page 689 and 690:
Author indexAboye, Teshome Leta 142
- Page 691 and 692:
Chi, Hongfang 480Chiche, Laurent 6C
- Page 693 and 694:
Geronikaki, Athina 444Gessmann, Ren
- Page 695 and 696:
Kostova, Kalina 528Kotzia, Georgia
- Page 697 and 698:
Nagata, Koji 162Nagayev, Igor Yu. 5
- Page 699 and 700:
Salvarese, Nicola 518Sammet, Benedi
- Page 701 and 702:
Vauquelin, Georges 138Veiga, Ana Sa