View - DSpace UniPR
View - DSpace UniPR
View - DSpace UniPR
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Chapter 5<br />
PNA 3 PNA 5 PNA 6<br />
Slide 1<br />
DNA c3 + DNA c5<br />
Slide 2<br />
DNA c3<br />
Slide 3<br />
DNA c5<br />
Slide 3 DNA c3<br />
60 bases<br />
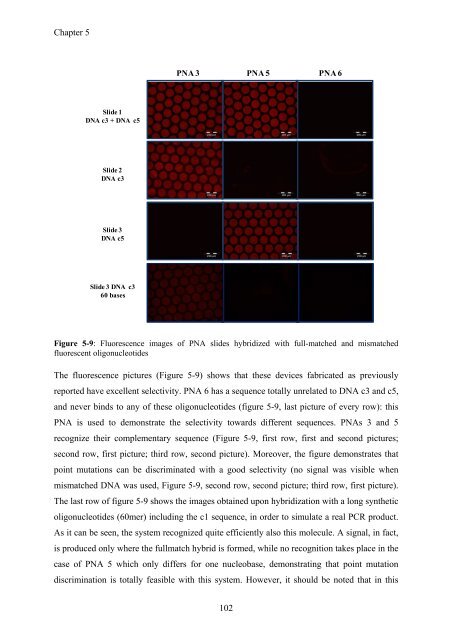
Figure 5-9: Fluorescence images of PNA slides hybridized with full-matched and mismatched<br />
fluorescent oligonucleotides<br />
The fluorescence pictures (Figure 5-9) shows that these devices fabricated as previously<br />
reported have excellent selectivity. PNA 6 has a sequence totally unrelated to DNA c3 and c5,<br />
and never binds to any of these oligonucleotides (figure 5-9, last picture of every row): this<br />
PNA is used to demonstrate the selectivity towards different sequences. PNAs 3 and 5<br />
recognize their complementary sequence (Figure 5-9, first row, first and second pictures;<br />
second row, first picture; third row, second picture). Moreover, the figure demonstrates that<br />
point mutations can be discriminated with a good selectivity (no signal was visible when<br />
mismatched DNA was used, Figure 5-9, second row, second picture; third row, first picture).<br />
The last row of figure 5-9 shows the images obtained upon hybridization with a long synthetic<br />
oligonucleotides (60mer) including the c1 sequence, in order to simulate a real PCR product.<br />
As it can be seen, the system recognized quite efficiently also this molecule. A signal, in fact,<br />
is produced only where the fullmatch hybrid is formed, while no recognition takes place in the<br />
case of PNA 5 which only differs for one nucleobase, demonstrating that point mutation<br />
discrimination is totally feasible with this system. However, it should be noted that in this<br />
102