View - DSpace UniPR
View - DSpace UniPR
View - DSpace UniPR
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
Chapter 4<br />
4.2.2 DNA recognition by PNA probes in solution<br />
Probe performances were initially tested in solution by measuring the melting temperatures of<br />
both fullmatch and mismatch PNA-DNA duplexes, by using oligonucleotides homologous to<br />
the ApoE sequences (Table 4-1). Melting temperatures were evaluated as the first derivatives<br />
of the UV absorption curves at 260 nm in a temperature range from 20 to 90°C.<br />
The chiral PNA probes were found to bind with very high stability to complementary DNA<br />
sequences (Tm from 69 to 85°C) and also exerted very high sequence recognition, with<br />
differences between full match and mismatch melting temperatures ranging from 15°C to<br />
19°C. The high stability of the full match complexes, together with the increased specificity,<br />
make feasible to find conditions, after PNA deposition on a microarray, in which it is possible<br />
to have good signals for full match duplexes and no signals for the mismatched ones.<br />
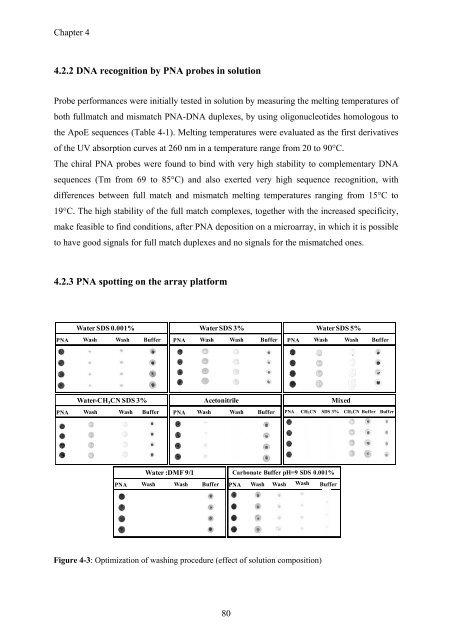
4.2.3 PNA spotting on the array platform<br />
Water SDS 0.001%<br />
PNA Wash Wash Buffer<br />
Water SDS 3%<br />
PNA Wash Wash Buffer<br />
Water SDS 5%<br />
PNA Wash Wash Buffer<br />
Water-CH 3 CN SDS 3%<br />
Acetonitrile<br />
Mixed<br />
PNA<br />
Wash<br />
Wash<br />
Buffer<br />
PNA Wash Wash Buffer<br />
PNA CH 3CN SDS 3% CH 3CN Buffer Buffer<br />
Water :DMF 9/1<br />
PNA Wash Wash Buffer<br />
Carbonate Buffer pH=9 SDS 0.001%<br />
PNA Wash Wash Wash Buffer<br />
Figure 4-3: Optimization of washing procedure (effect of solution composition)<br />
80