View - DSpace UniPR
View - DSpace UniPR
View - DSpace UniPR
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
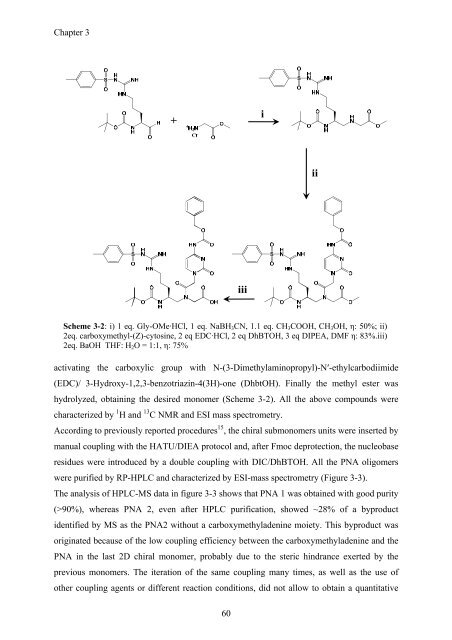
Chapter 3<br />
+<br />
i<br />
ii<br />
iii<br />
Scheme 3-2: i) 1 eq. Gly-OMe·HCl, 1 eq. NaBH 3 CN, 1.1 eq. CH 3 COOH, CH 3 OH, η: 50%; ii)<br />
2eq. carboxymethyl-(Z)-cytosine, 2 eq EDC·HCl, 2 eq DhBTOH, 3 eq DIPEA, DMF η: 83%.iii)<br />
2eq. BaOH THF: H 2 O = 1:1, η: 75%<br />
activating the carboxylic group with N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide<br />
(EDC)/ 3-Hydroxy-1,2,3-benzotriazin-4(3H)-one (DhbtOH). Finally the methyl ester was<br />
hydrolyzed, obtaining the desired monomer (Scheme 3-2). All the above compounds were<br />
characterized by 1 H and 13 C NMR and ESI mass spectrometry.<br />
According to previously reported procedures 15 , the chiral submonomers units were inserted by<br />
manual coupling with the HATU/DIEA protocol and, after Fmoc deprotection, the nucleobase<br />
residues were introduced by a double coupling with DIC/DhBTOH. All the PNA oligomers<br />
were purified by RP-HPLC and characterized by ESI-mass spectrometry (Figure 3-3).<br />
The analysis of HPLC-MS data in figure 3-3 shows that PNA 1 was obtained with good purity<br />
(>90%), whereas PNA 2, even after HPLC purification, showed ~28% of a byproduct<br />
identified by MS as the PNA2 without a carboxymethyladenine moiety. This byproduct was<br />
originated because of the low coupling efficiency between the carboxymethyladenine and the<br />
PNA in the last 2D chiral monomer, probably due to the steric hindrance exerted by the<br />
previous monomers. The iteration of the same coupling many times, as well as the use of<br />
other coupling agents or different reaction conditions, did not allow to obtain a quantitative<br />
60