View - DSpace UniPR
View - DSpace UniPR
View - DSpace UniPR
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
Chapter 3<br />
with those of the analogous PNA bearing in the middle a chiral monomer 2D,5L based on<br />
lysine.<br />
Moreover, since the introduction of several adjacent positively charged monomers in the<br />
sequence was demonstrated to improve the binding selectivity, another PNA was designed<br />
combining the features of the “chiral box” PNA and those of 2D,5L PNA. This new PNA was<br />
designed with a 2D,5L arginine-based monomer in the middle, which should determine an<br />
enhanced affinity for the complementary DNA sequence, flanked by 2D-modified and a 5Lmodified<br />
arginine-based monomers, which should improve the PNA selectivity in mismatch<br />
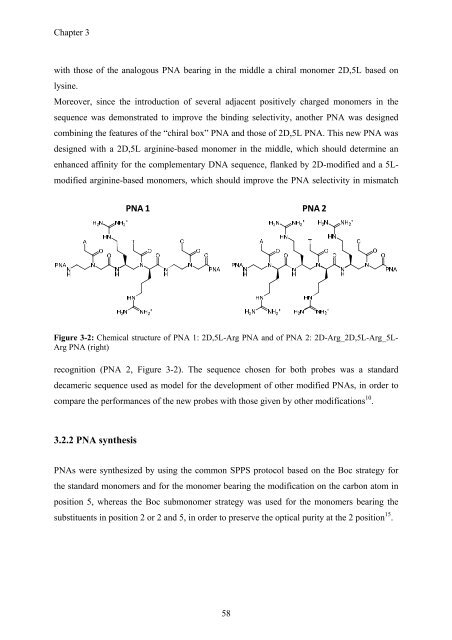
PNA 1 PNA 2<br />
Figure 3-2: Chemical structure of PNA 1: 2D,5L-Arg PNA and of PNA 2: 2D-Arg_2D,5L-Arg_5L-<br />
Arg PNA (right)<br />
recognition (PNA 2, Figure 3-2). The sequence chosen for both probes was a standard<br />
decameric sequence used as model for the development of other modified PNAs, in order to<br />
compare the performances of the new probes with those given by other modifications 10 .<br />
3.2.2 PNA synthesis<br />
PNAs were synthesized by using the common SPPS protocol based on the Boc strategy for<br />
the standard monomers and for the monomer bearing the modification on the carbon atom in<br />
position 5, whereas the Boc submonomer strategy was used for the monomers bearing the<br />
substituents in position 2 or 2 and 5, in order to preserve the optical purity at the 2 position 15 .<br />
58