View - DSpace UniPR
View - DSpace UniPR
View - DSpace UniPR
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
Chapter 2<br />
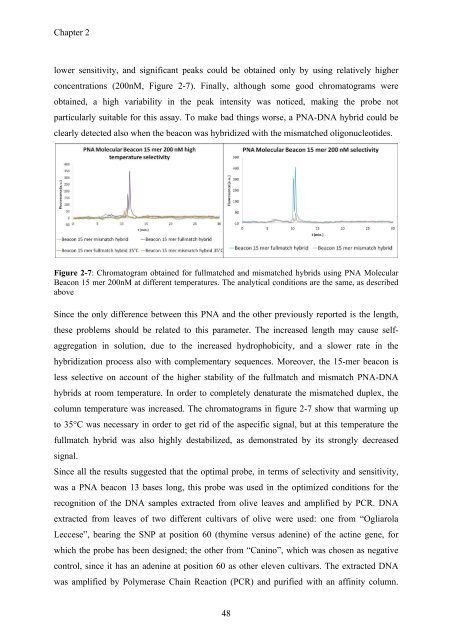
lower sensitivity, and significant peaks could be obtained only by using relatively higher<br />
concentrations (200nM, Figure 2-7). Finally, although some good chromatograms were<br />
obtained, a high variability in the peak intensity was noticed, making the probe not<br />
particularly suitable for this assay. To make bad things worse, a PNA-DNA hybrid could be<br />
clearly detected also when the beacon was hybridized with the mismatched oligonucleotides.<br />
Figure 2-7: Chromatogram obtained for fullmatched and mismatched hybrids using PNA Molecular<br />
Beacon 15 mer 200nM at different temperatures. The analytical conditions are the same, as described<br />
above<br />
Since the only difference between this PNA and the other previously reported is the length,<br />
these problems should be related to this parameter. The increased length may cause selfaggregation<br />
in solution, due to the increased hydrophobicity, and a slower rate in the<br />
hybridization process also with complementary sequences. Moreover, the 15-mer beacon is<br />
less selective on account of the higher stability of the fullmatch and mismatch PNA-DNA<br />
hybrids at room temperature. In order to completely denaturate the mismatched duplex, the<br />
column temperature was increased. The chromatograms in figure 2-7 show that warming up<br />
to 35°C was necessary in order to get rid of the aspecific signal, but at this temperature the<br />
fullmatch hybrid was also highly destabilized, as demonstrated by its strongly decreased<br />
signal.<br />
Since all the results suggested that the optimal probe, in terms of selectivity and sensitivity,<br />
was a PNA beacon 13 bases long, this probe was used in the optimized conditions for the<br />
recognition of the DNA samples extracted from olive leaves and amplified by PCR. DNA<br />
extracted from leaves of two different cultivars of olive were used: one from “Ogliarola<br />
Leccese”, bearing the SNP at position 60 (thymine versus adenine) of the actine gene, for<br />
which the probe has been designed; the other from “Canino”, which was chosen as negative<br />
control, since it has an adenine at position 60 as other eleven cultivars. The extracted DNA<br />
was amplified by Polymerase Chain Reaction (PCR) and purified with an affinity column.<br />
48