Gene regulation in Streptococcus pneumoniae - RePub - Erasmus ...
Gene regulation in Streptococcus pneumoniae - RePub - Erasmus ...
Gene regulation in Streptococcus pneumoniae - RePub - Erasmus ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
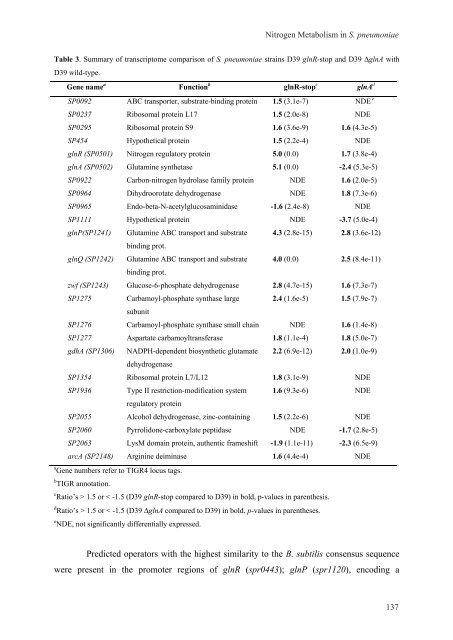
Table 3. Summary of transcriptome comparison of S. <strong>pneumoniae</strong> stra<strong>in</strong>s D39 glnR-stop and D39 ΔglnA with<br />
D39 wild-type.<br />
<strong>Gene</strong> name a Function b glnR-stop c glnA d<br />
SP0092 ABC transporter, substrate-b<strong>in</strong>d<strong>in</strong>g prote<strong>in</strong> 1.5 (3.1e-7) NDE e<br />
SP0237 Ribosomal prote<strong>in</strong> L17 1.5 (2.0e-8) NDE<br />
SP0295 Ribosomal prote<strong>in</strong> S9 1.6 (3.6e-9) 1.6 (4.3e-5)<br />
SP454 Hypothetical prote<strong>in</strong> 1.5 (2.2e-4) NDE<br />
glnR (SP0501) Nitrogen regulatory prote<strong>in</strong> 5.0 (0.0) 1.7 (3.8e-4)<br />
glnA (SP0502) Glutam<strong>in</strong>e synthetase 5.1 (0.0) -2.4 (5.3e-5)<br />
SP0922 Carbon-nitrogen hydrolase family prote<strong>in</strong> NDE 1.6 (2.0e-5)<br />
SP0964 Dihydroorotate dehydrogenase NDE 1.8 (7.3e-6)<br />
SP0965 Endo-beta-N-acetylglucosam<strong>in</strong>idase -1.6 (2.4e-8) NDE<br />
SP1111 Hypothetical prote<strong>in</strong> NDE -3.7 (5.0e-4)<br />
glnP(SP1241) Glutam<strong>in</strong>e ABC transport and substrate<br />
b<strong>in</strong>d<strong>in</strong>g prot.<br />
4.3 (2.8e-15) 2.8 (3.6e-12)<br />
glnQ (SP1242) Glutam<strong>in</strong>e ABC transport and substrate<br />
b<strong>in</strong>d<strong>in</strong>g prot.<br />
4.0 (0.0) 2.5 (8.4e-11)<br />
zwf (SP1243) Glucose-6-phosphate dehydrogenase 2.8 (4.7e-15) 1.6 (7.3e-7)<br />
SP1275 Carbamoyl-phosphate synthase large<br />
subunit<br />
2.4 (1.6e-5) 1.5 (7.9e-7)<br />
SP1276 Carbamoyl-phosphate synthase small cha<strong>in</strong> NDE 1.6 (1.4e-8)<br />
SP1277 Aspartate carbamoyltransferase 1.8 (1.1e-4) 1.8 (5.0e-7)<br />
gdhA (SP1306) NADPH-dependent biosynthetic glutamate<br />
dehydrogenase<br />
2.2 (6.9e-12) 2.0 (1.0e-9)<br />
SP1354 Ribosomal prote<strong>in</strong> L7/L12 1.8 (3.1e-9) NDE<br />
SP1936 Type II restriction-modification system<br />
regulatory prote<strong>in</strong><br />
1.6 (9.3e-6) NDE<br />
SP2055 Alcohol dehydrogenase, z<strong>in</strong>c-conta<strong>in</strong><strong>in</strong>g 1.5 (2.2e-6) NDE<br />
SP2060 Pyrrolidone-carboxylate peptidase NDE -1.7 (2.8e-5)<br />
SP2063 LysM doma<strong>in</strong> prote<strong>in</strong>, authentic frameshift -1.9 (1.1e-11) -2.3 (6.5e-9)<br />
arcA (SP2148) Arg<strong>in</strong><strong>in</strong>e deim<strong>in</strong>ase 1.6 (4.4e-4) NDE<br />
a<br />
<strong>Gene</strong> numbers refer to TIGR4 locus tags.<br />
b<br />
TIGR annotation.<br />
c<br />
Ratio’s > 1.5 or < -1.5 (D39 glnR-stop compared to D39) <strong>in</strong> bold, p-values <strong>in</strong> parenthesis.<br />
d Ratio’s > 1.5 or < -1.5 (D39 ΔglnA compared to D39) <strong>in</strong> bold, p-values <strong>in</strong> parentheses.<br />
e NDE, not significantly differentially expressed.<br />
Nitrogen Metabolism <strong>in</strong> S. <strong>pneumoniae</strong><br />
Predicted operators with the highest similarity to the B. subtilis consensus sequence<br />
were present <strong>in</strong> the promoter regions of glnR (spr0443); glnP (spr1120), encod<strong>in</strong>g a<br />
137<br />
137