Gene regulation in Streptococcus pneumoniae - RePub - Erasmus ...
Gene regulation in Streptococcus pneumoniae - RePub - Erasmus ...
Gene regulation in Streptococcus pneumoniae - RePub - Erasmus ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
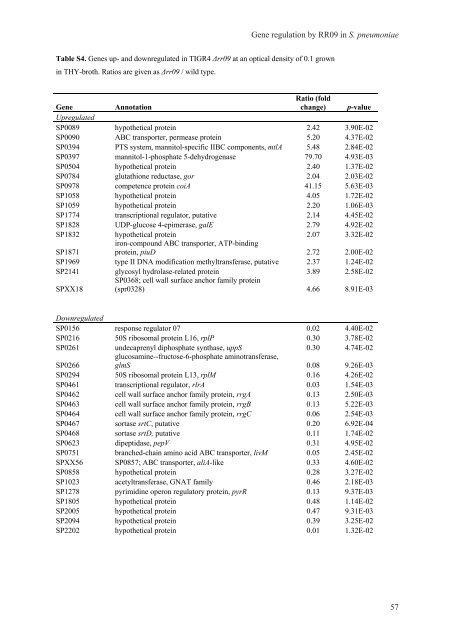
Table S4. <strong>Gene</strong>s up- and downregulated <strong>in</strong> TIGR4 Δrr09 at an optical density of 0.1 grown<br />
<strong>in</strong> THY-broth. Ratios are given as Δrr09 / wild type.<br />
<strong>Gene</strong> <strong>regulation</strong> by RR09 <strong>in</strong> S. <strong>pneumoniae</strong><br />
<strong>Gene</strong><br />
Upregulated<br />
Annotation<br />
Ratio (fold<br />
change) p-value<br />
SP0089 hypothetical prote<strong>in</strong> 2.42 3.90E-02<br />
SP0090 ABC transporter, permease prote<strong>in</strong> 5.20 4.37E-02<br />
SP0394 PTS system, mannitol-specific IIBC components, mtlA 5.48 2.84E-02<br />
SP0397 mannitol-1-phosphate 5-dehydrogenase 79.70 4.93E-03<br />
SP0504 hypothetical prote<strong>in</strong> 2.40 1.37E-02<br />
SP0784 glutathione reductase, gor 2.04 2.03E-02<br />
SP0978 competence prote<strong>in</strong> coiA 41.15 5.63E-03<br />
SP1058 hypothetical prote<strong>in</strong> 4.05 1.72E-02<br />
SP1059 hypothetical prote<strong>in</strong> 2.20 1.06E-03<br />
SP1774 transcriptional regulator, putative 2.14 4.45E-02<br />
SP1828 UDP-glucose 4-epimerase, galE 2.79 4.92E-02<br />
SP1832 hypothetical prote<strong>in</strong><br />
iron-compound ABC transporter, ATP-b<strong>in</strong>d<strong>in</strong>g<br />
2.07 3.32E-02<br />
SP1871 prote<strong>in</strong>, piuD 2.72 2.00E-02<br />
SP1969 type II DNA modification methyltransferase, putative 2.37 1.24E-02<br />
SP2141 glycosyl hydrolase-related prote<strong>in</strong><br />
SP0368; cell wall surface anchor family prote<strong>in</strong><br />
3.89 2.58E-02<br />
SPXX18 (spr0328) 4.66 8.91E-03<br />
Downregulated<br />
SP0156 response regulator 07 0.02 4.40E-02<br />
SP0216 50S ribosomal prote<strong>in</strong> L16, rplP 0.30 3.78E-02<br />
SP0261 undecaprenyl diphosphate synthase, uppS<br />
glucosam<strong>in</strong>e--fructose-6-phosphate am<strong>in</strong>otransferase,<br />
0.30 4.74E-02<br />
SP0266 glmS 0.08 9.26E-03<br />
SP0294 50S ribosomal prote<strong>in</strong> L13, rplM 0.16 4.26E-02<br />
SP0461 transcriptional regulator, rlrA 0.03 1.54E-03<br />
SP0462 cell wall surface anchor family prote<strong>in</strong>, rrgA 0.13 2.50E-03<br />
SP0463 cell wall surface anchor family prote<strong>in</strong>, rrgB 0.13 5.22E-03<br />
SP0464 cell wall surface anchor family prote<strong>in</strong>, rrgC 0.06 2.54E-03<br />
SP0467 sortase srtC, putative 0.20 6.92E-04<br />
SP0468 sortase srtD, putative 0.11 1.74E-02<br />
SP0623 dipeptidase, pepV 0.31 4.95E-02<br />
SP0751 branched-cha<strong>in</strong> am<strong>in</strong>o acid ABC transporter, livM 0.05 2.45E-02<br />
SPXX56 SP0857; ABC transporter, aliA-like 0.33 4.60E-02<br />
SP0858 hypothetical prote<strong>in</strong> 0.28 3.27E-02<br />
SP1023 acetyltransferase, GNAT family 0.46 2.18E-03<br />
SP1278 pyrimid<strong>in</strong>e operon regulatory prote<strong>in</strong>, pyrR 0.13 9.37E-03<br />
SP1805 hypothetical prote<strong>in</strong> 0.48 1.14E-02<br />
SP2005 hypothetical prote<strong>in</strong> 0.47 9.31E-03<br />
SP2094 hypothetical prote<strong>in</strong> 0.39 3.25E-02<br />
SP2202 hypothetical prote<strong>in</strong> 0.01 1.32E-02<br />
57<br />
57