Gene regulation in Streptococcus pneumoniae - RePub - Erasmus ...
Gene regulation in Streptococcus pneumoniae - RePub - Erasmus ...
Gene regulation in Streptococcus pneumoniae - RePub - Erasmus ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Chapter 2<br />
Supplemental material<br />
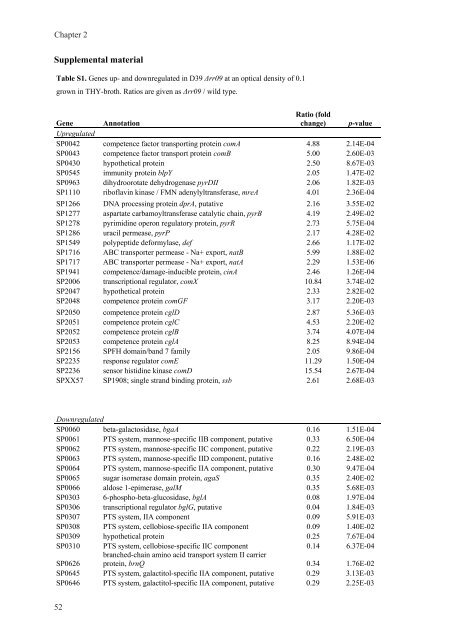
Table S1. <strong>Gene</strong>s up- and downregulated <strong>in</strong> D39 Δrr09 at an optical density of 0.1<br />
grown <strong>in</strong> THY-broth. Ratios are given as Δrr09 / wild type.<br />
<strong>Gene</strong><br />
Upregulated<br />
Annotation<br />
Ratio (fold<br />
change) p-value<br />
SP0042 competence factor transport<strong>in</strong>g prote<strong>in</strong> comA 4.88 2.14E-04<br />
SP0043 competence factor transport prote<strong>in</strong> comB 5.00 2.60E-03<br />
SP0430 hypothetical prote<strong>in</strong> 2.50 8.67E-03<br />
SP0545 immunity prote<strong>in</strong> blpY 2.05 1.47E-02<br />
SP0963 dihydroorotate dehydrogenase pyrDII 2.06 1.82E-03<br />
SP1110 riboflav<strong>in</strong> k<strong>in</strong>ase / FMN adenylyltransferase, mreA 4.01 2.36E-04<br />
SP1266 DNA process<strong>in</strong>g prote<strong>in</strong> dprA, putative 2.16 3.55E-02<br />
SP1277 aspartate carbamoyltransferase catalytic cha<strong>in</strong>, pyrB 4.19 2.49E-02<br />
SP1278 pyrimid<strong>in</strong>e operon regulatory prote<strong>in</strong>, pyrR 2.73 5.75E-04<br />
SP1286 uracil permease, pyrP 2.17 4.28E-02<br />
SP1549 polypeptide deformylase, def 2.66 1.17E-02<br />
SP1716 ABC transporter permease - Na+ export, natB 5.99 1.88E-02<br />
SP1717 ABC transporter permease - Na+ export, natA 2.29 1.53E-06<br />
SP1941 competence/damage-<strong>in</strong>ducible prote<strong>in</strong>, c<strong>in</strong>A 2.46 1.26E-04<br />
SP2006 transcriptional regulator, comX 10.84 3.74E-02<br />
SP2047 hypothetical prote<strong>in</strong> 2.33 2.82E-02<br />
SP2048 competence prote<strong>in</strong> comGF 3.17 2.20E-03<br />
SP2050 competence prote<strong>in</strong> cglD 2.87 5.36E-03<br />
SP2051 competence prote<strong>in</strong> cglC 4.53 2.20E-02<br />
SP2052 competence prote<strong>in</strong> cglB 3.74 4.07E-04<br />
SP2053 competence prote<strong>in</strong> cglA 8.25 8.94E-04<br />
SP2156 SPFH doma<strong>in</strong>/band 7 family 2.05 9.86E-04<br />
SP2235 response regulator comE 11.29 1.50E-04<br />
SP2236 sensor histid<strong>in</strong>e k<strong>in</strong>ase comD 15.54 2.67E-04<br />
SPXX57 SP1908; s<strong>in</strong>gle strand b<strong>in</strong>d<strong>in</strong>g prote<strong>in</strong>, ssb 2.61 2.68E-03<br />
Downregulated<br />
SP0060 beta-galactosidase, bgaA 0.16 1.51E-04<br />
SP0061 PTS system, mannose-specific IIB component, putative 0.33 6.50E-04<br />
SP0062 PTS system, mannose-specific IIC component, putative 0.22 2.19E-03<br />
SP0063 PTS system, mannose-specific IID component, putative 0.16 2.48E-02<br />
SP0064 PTS system, mannose-specific IIA component, putative 0.30 9.47E-04<br />
SP0065 sugar isomerase doma<strong>in</strong> prote<strong>in</strong>, agaS 0.35 2.40E-02<br />
SP0066 aldose 1-epimerase, galM 0.35 5.68E-03<br />
SP0303 6-phospho-beta-glucosidase, bglA 0.08 1.97E-04<br />
SP0306 transcriptional regulator bglG, putative 0.04 1.84E-03<br />
SP0307 PTS system, IIA component 0.09 5.91E-03<br />
SP0308 PTS system, cellobiose-specific IIA component 0.09 1.40E-02<br />
SP0309 hypothetical prote<strong>in</strong> 0.25 7.67E-04<br />
SP0310 PTS system, cellobiose-specific IIC component<br />
branched-cha<strong>in</strong> am<strong>in</strong>o acid transport system II carrier<br />
0.14 6.37E-04<br />
SP0626 prote<strong>in</strong>, brnQ 0.34 1.76E-02<br />
SP0645 PTS system, galactitol-specific IIA component, putative 0.29 3.13E-03<br />
SP0646 PTS system, galactitol-specific IIA component, putative 0.29 2.25E-03<br />
52<br />
52