Gene regulation in Streptococcus pneumoniae - RePub - Erasmus ...
Gene regulation in Streptococcus pneumoniae - RePub - Erasmus ...
Gene regulation in Streptococcus pneumoniae - RePub - Erasmus ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
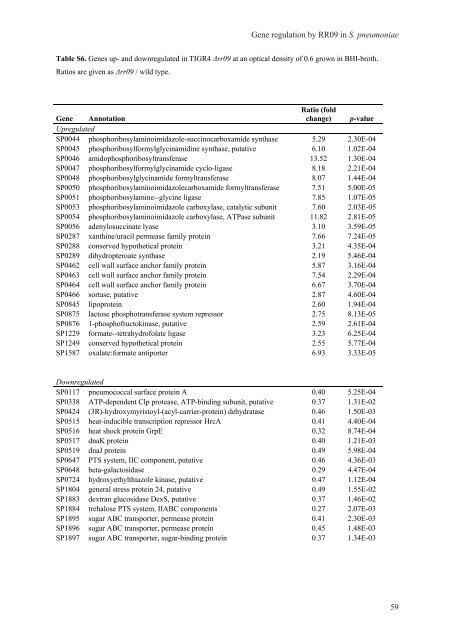
Table S6. <strong>Gene</strong>s up- and downregulated <strong>in</strong> TIGR4 Δrr09 at an optical density of 0.6 grown <strong>in</strong> BHI-broth.<br />
Ratios are given as Δrr09 / wild type.<br />
<strong>Gene</strong> <strong>regulation</strong> by RR09 <strong>in</strong> S. <strong>pneumoniae</strong><br />
<strong>Gene</strong> Annotation<br />
Ratio (fold<br />
change) p-value<br />
Upregulated<br />
SP0044 phosphoribosylam<strong>in</strong>oimidazole-succ<strong>in</strong>ocarboxamide synthase 5.29 2.30E-04<br />
SP0045 phosphoribosylformylglyc<strong>in</strong>amid<strong>in</strong>e synthase, putative 6.10 1.02E-04<br />
SP0046 amidophosphoribosyltransferase 13.52 1.30E-04<br />
SP0047 phosphoribosylformylglyc<strong>in</strong>amide cyclo-ligase 8.18 2.21E-04<br />
SP0048 phosphoribosylglyc<strong>in</strong>amide formyltransferase 8.07 1.44E-04<br />
SP0050 phosphoribosylam<strong>in</strong>oimidazolecarboxamide formyltransferase 7.51 5.00E-05<br />
SP0051 phosphoribosylam<strong>in</strong>e--glyc<strong>in</strong>e ligase 7.85 1.07E-05<br />
SP0053 phosphoribosylam<strong>in</strong>oimidazole carboxylase, catalytic subunit 7.60 2.03E-05<br />
SP0054 phosphoribosylam<strong>in</strong>oimidazole carboxylase, ATPase subunit 11.82 2.81E-05<br />
SP0056 adenylosucc<strong>in</strong>ate lyase 3.10 3.59E-05<br />
SP0287 xanth<strong>in</strong>e/uracil permease family prote<strong>in</strong> 7.66 7.24E-05<br />
SP0288 conserved hypothetical prote<strong>in</strong> 3.21 4.35E-04<br />
SP0289 dihydropteroate synthase 2.19 5.46E-04<br />
SP0462 cell wall surface anchor family prote<strong>in</strong> 5.87 3.16E-04<br />
SP0463 cell wall surface anchor family prote<strong>in</strong> 7.54 2.29E-04<br />
SP0464 cell wall surface anchor family prote<strong>in</strong> 6.67 3.70E-04<br />
SP0466 sortase, putative 2.87 4.60E-04<br />
SP0845 lipoprote<strong>in</strong> 2.60 1.94E-04<br />
SP0875 lactose phosphotransferase system repressor 2.75 8.13E-05<br />
SP0876 1-phosphofructok<strong>in</strong>ase, putative 2.59 2.61E-04<br />
SP1229 formate--tetrahydrofolate ligase 3.23 6.25E-04<br />
SP1249 conserved hypothetical prote<strong>in</strong> 2.55 5.77E-04<br />
SP1587 oxalate:formate antiporter 6.93 3.33E-05<br />
Downregulated<br />
SP0117 pneumococcal surface prote<strong>in</strong> A 0.40 5.25E-04<br />
SP0338 ATP-dependent Clp protease, ATP-b<strong>in</strong>d<strong>in</strong>g subunit, putative 0.37 1.31E-02<br />
SP0424 (3R)-hydroxymyristoyl-(acyl-carrier-prote<strong>in</strong>) dehydratase 0.46 1.50E-03<br />
SP0515 heat-<strong>in</strong>ducible transcription repressor HrcA 0.41 4.40E-04<br />
SP0516 heat shock prote<strong>in</strong> GrpE 0.32 8.74E-04<br />
SP0517 dnaK prote<strong>in</strong> 0.40 1.21E-03<br />
SP0519 dnaJ prote<strong>in</strong> 0.49 5.98E-04<br />
SP0647 PTS system, IIC component, putative 0.46 4.36E-03<br />
SP0648 beta-galactosidase 0.29 4.47E-04<br />
SP0724 hydroxyethylthiazole k<strong>in</strong>ase, putative 0.47 1.12E-04<br />
SP1804 general stress prote<strong>in</strong> 24, putative 0.49 1.55E-02<br />
SP1883 dextran glucosidase DexS, putative 0.37 1.46E-02<br />
SP1884 trehalose PTS system, IIABC components 0.27 2.07E-03<br />
SP1895 sugar ABC transporter, permease prote<strong>in</strong> 0.41 2.30E-03<br />
SP1896 sugar ABC transporter, permease prote<strong>in</strong> 0.45 1.48E-03<br />
SP1897 sugar ABC transporter, sugar-b<strong>in</strong>d<strong>in</strong>g prote<strong>in</strong> 0.37 1.34E-03<br />
59<br />
59