Gene regulation in Streptococcus pneumoniae - RePub - Erasmus ...
Gene regulation in Streptococcus pneumoniae - RePub - Erasmus ...
Gene regulation in Streptococcus pneumoniae - RePub - Erasmus ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Chapter 2<br />
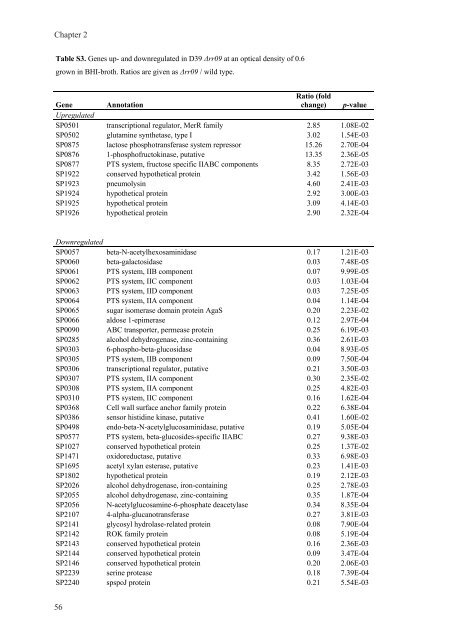
Table S3. <strong>Gene</strong>s up- and downregulated <strong>in</strong> D39 Δrr09 at an optical density of 0.6<br />
grown <strong>in</strong> BHI-broth. Ratios are given as Δrr09 / wild type.<br />
<strong>Gene</strong><br />
Upregulated<br />
Annotation<br />
Ratio (fold<br />
change) p-value<br />
SP0501 transcriptional regulator, MerR family 2.85 1.08E-02<br />
SP0502 glutam<strong>in</strong>e synthetase, type I 3.02 1.54E-03<br />
SP0875 lactose phosphotransferase system repressor 15.26 2.70E-04<br />
SP0876 1-phosphofructok<strong>in</strong>ase, putative 13.35 2.36E-05<br />
SP0877 PTS system, fructose specific IIABC components 8.35 2.72E-03<br />
SP1922 conserved hypothetical prote<strong>in</strong> 3.42 1.56E-03<br />
SP1923 pneumolys<strong>in</strong> 4.60 2.41E-03<br />
SP1924 hypothetical prote<strong>in</strong> 2.92 3.00E-03<br />
SP1925 hypothetical prote<strong>in</strong> 3.09 4.14E-03<br />
SP1926 hypothetical prote<strong>in</strong> 2.90 2.32E-04<br />
Downregulated<br />
SP0057 beta-N-acetylhexosam<strong>in</strong>idase 0.17 1.21E-03<br />
SP0060 beta-galactosidase 0.03 7.48E-05<br />
SP0061 PTS system, IIB component 0.07 9.99E-05<br />
SP0062 PTS system, IIC component 0.03 1.03E-04<br />
SP0063 PTS system, IID component 0.03 7.25E-05<br />
SP0064 PTS system, IIA component 0.04 1.14E-04<br />
SP0065 sugar isomerase doma<strong>in</strong> prote<strong>in</strong> AgaS 0.20 2.23E-02<br />
SP0066 aldose 1-epimerase 0.12 2.97E-04<br />
SP0090 ABC transporter, permease prote<strong>in</strong> 0.25 6.19E-03<br />
SP0285 alcohol dehydrogenase, z<strong>in</strong>c-conta<strong>in</strong><strong>in</strong>g 0.36 2.61E-03<br />
SP0303 6-phospho-beta-glucosidase 0.04 8.93E-05<br />
SP0305 PTS system, IIB component 0.09 7.50E-04<br />
SP0306 transcriptional regulator, putative 0.21 3.50E-03<br />
SP0307 PTS system, IIA component 0.30 2.35E-02<br />
SP0308 PTS system, IIA component 0.25 4.82E-03<br />
SP0310 PTS system, IIC component 0.16 1.62E-04<br />
SP0368 Cell wall surface anchor family prote<strong>in</strong> 0.22 6.38E-04<br />
SP0386 sensor histid<strong>in</strong>e k<strong>in</strong>ase, putative 0.41 1.60E-02<br />
SP0498 endo-beta-N-acetylglucosam<strong>in</strong>idase, putative 0.19 5.05E-04<br />
SP0577 PTS system, beta-glucosides-specific IIABC 0.27 9.38E-03<br />
SP1027 conserved hypothetical prote<strong>in</strong> 0.25 1.37E-02<br />
SP1471 oxidoreductase, putative 0.33 6.98E-03<br />
SP1695 acetyl xylan esterase, putative 0.23 1.41E-03<br />
SP1802 hypothetical prote<strong>in</strong> 0.19 2.12E-03<br />
SP2026 alcohol dehydrogenase, iron-conta<strong>in</strong><strong>in</strong>g 0.25 2.78E-03<br />
SP2055 alcohol dehydrogenase, z<strong>in</strong>c-conta<strong>in</strong><strong>in</strong>g 0.35 1.87E-04<br />
SP2056 N-acetylglucosam<strong>in</strong>e-6-phosphate deacetylase 0.34 8.35E-04<br />
SP2107 4-alpha-glucanotransferase 0.27 3.81E-03<br />
SP2141 glycosyl hydrolase-related prote<strong>in</strong> 0.08 7.90E-04<br />
SP2142 ROK family prote<strong>in</strong> 0.08 5.19E-04<br />
SP2143 conserved hypothetical prote<strong>in</strong> 0.16 2.36E-03<br />
SP2144 conserved hypothetical prote<strong>in</strong> 0.09 3.47E-04<br />
SP2146 conserved hypothetical prote<strong>in</strong> 0.20 2.06E-03<br />
SP2239 ser<strong>in</strong>e protease 0.18 7.39E-04<br />
SP2240 spspoJ prote<strong>in</strong> 0.21 5.54E-03<br />
56<br />
56