Gene regulation in Streptococcus pneumoniae - RePub - Erasmus ...
Gene regulation in Streptococcus pneumoniae - RePub - Erasmus ...
Gene regulation in Streptococcus pneumoniae - RePub - Erasmus ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
adjacent genes, encod<strong>in</strong>g a MerR family transcriptional regulator (sp1856) and czcD<br />
(sp1857), a Zn 2+ -efflux pump (Table 3).<br />
Four genes were downregulated: guaC, encod<strong>in</strong>g GMP reductase (sp1249), ply,<br />
encod<strong>in</strong>g pneumolys<strong>in</strong> (sp1923), and two genes encod<strong>in</strong>g the B and C component of a<br />
putative cellobiose-specific PTS (sp2022 and sp2023). The complete set of differentially<br />
expressed genes <strong>in</strong> TIGR4∆psaR is given <strong>in</strong> Table 3.<br />
Proteome analysis of ΔpsaR <strong>in</strong> D39 and TIGR4<br />
To exam<strong>in</strong>e if the observed PsaR-mediated differences <strong>in</strong> gene expression<br />
corresponded with changes <strong>in</strong> prote<strong>in</strong> expression, we performed SILAC for both wild-type<br />
stra<strong>in</strong>s and their psaR mutants. To this end, wild-type and isogenic ΔpsaR counterparts were<br />
cultured <strong>in</strong> the presence of stable isotope-labeled (heavy, 13 C6) or normal (light, 12 C6) L-lys<strong>in</strong>e<br />
and L-arg<strong>in</strong><strong>in</strong>e, respectively. Equal amounts of bacteria were mixed and analyzed by mass<br />
spectrometry, after which changes <strong>in</strong> prote<strong>in</strong> expression between wild-type and isogenic<br />
ΔpsaR stra<strong>in</strong>s were derived from the correspond<strong>in</strong>g heavy to light peptide ratios.<br />
In D39ΔpsaR, n<strong>in</strong>e prote<strong>in</strong>s were found to be more abundant than <strong>in</strong> the wild-type<br />
(Table 3). Most pronounced were FtsY, PsaA, SufS, Cps2G, CysK, and PrtA, all of which<br />
displayed expression levels at least eight times higher <strong>in</strong> the psaR-mutant than <strong>in</strong> the wildtype.<br />
No prote<strong>in</strong>s were identified that were less abundant <strong>in</strong> D39ΔpsaR (Table 3).<br />
In TIGR4ΔpsaR, six prote<strong>in</strong>s were more abundant than <strong>in</strong> the wild-type. Most<br />
pronounced were PcpA, PsaA, and RrgB, all with at least a four-fold <strong>in</strong>crease <strong>in</strong> expression <strong>in</strong><br />
the psaR-mutant (Table 3). Three prote<strong>in</strong>s were less abundant <strong>in</strong> TIGR4ΔpsaR, i.e., an ABCtransporter<br />
of a putative bacterioc<strong>in</strong> system (sp0148), TrxB, and ZmpB (Table 3).<br />
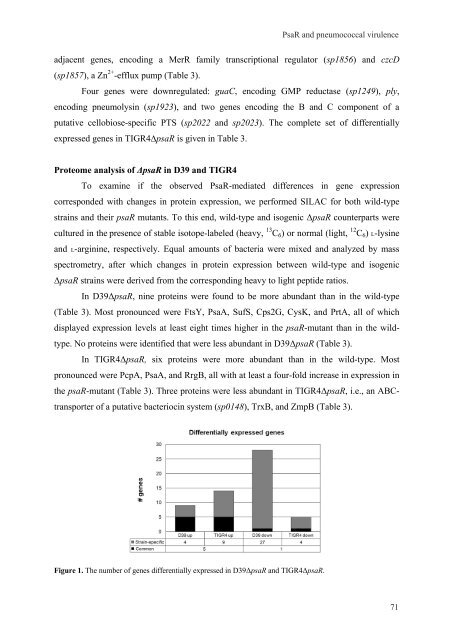
Figure 1. The number of genes differentially expressed <strong>in</strong> D39ΔpsaR and TIGR4ΔpsaR.<br />
PsaR and pneumococcal virulence<br />
71<br />
71