Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
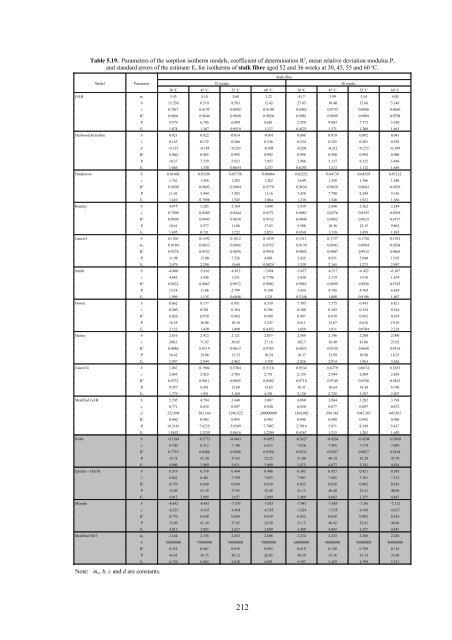
Table 5.19. Parameters of the sorption isotherm models, coefficient of determination R 2 , mean relative deviation modulus P,<br />
and standard errors of the estimate E s for isotherms of stalk fibre aged 52 and 36 weeks at 30, 45, 55 and 60 o C.<br />
Stalk fibre<br />
Model Parameter 52 weeks 36 weeks<br />
30 o C 45 o C 55 o C 60 o C 30 o C 45 o C 55 o C 60 o C<br />
GAB mo 5.43 5.18 3.68 3.22 4.17 3.99 3.54 4.00<br />
b 15.350 6.519 8.583 12.42 27.85 10.40 12.66 5.146<br />
c 0.7817 0.8193 0.8892 0.9140 0.8462 0.8733 0.8886 0.8660<br />
R 2 0.9601 0.9846 0.9949 0.9920 0.9981 0.9899 0.9909 0.9798<br />
P 9.579 6.705 6.099 8.681 2.359 9.083 7.773 5.550<br />
Es 1.874 1.367 0.9519 1.337 0.4523 3.371 1.208 1.663<br />
Hailwood Horrobin b 0.021 0.022 0.014 -0.001 0.006 0.019 0.002 0.041<br />
c 0.143 0.172 0.266 0.336 0.234 0.225 0.301 0.192<br />
d -0.123 -0.158 -0.250 -0.308 -0.204 -0.212 -0.272 -0.199<br />
R 2 0.960 0.985 0.995 0.992 0.998 0.990 0.992 0.980<br />
P 10.37 7.759 5.923 7.857 2.986 5.137 8.152 5.460<br />
Es 1.866 1.330 0.8654 1.257 0.6292 1.213 1.112 1.656<br />
Henderson b 0.01600 0.03501 0.05738 0.06064 0.02252 0.04735 0.04559 0.07212<br />
c 1.761 1.456 1.292 1.262 1.649 1.358 1.386 1.240<br />
R 2 0.9590 0.9893 0.9904 0.9779 0.9830 0.9859 0.9843 0.9939<br />
P 11.01 5.444 7.283 11.16 7.458 7.790 8.294 5.156<br />
Es 1.610 0.7998 1.545 3.004 1.239 1.548 1.923 1.656<br />
Bradley b 4.975 3.285 2.164 1.890 3.539 2.448 2.362 2.244<br />
c 0.7909 0.8209 0.8444 0.8571 0.8085 0.8376 0.8355 0.8309<br />
R 2 0.9590 0.9943 0.9878 0.9732 0.9948 0.9852 0.9815 0.9737<br />
P 10.61 5.577 11.86 17.83 5.588 10.36 12.15 9.862<br />
Es 1.695 0.731 1.252 2.053 0.6541 1.318 1.458 1.693<br />
Caurie I b 0.1401 0.1592 0.1812 0.1839 0.1511 0.1737 0.1720 0.1923<br />
mo 0.9180 0.8833 0.8865 0.8752 0.9139 0.8843 0.8984 0.9204<br />
R 2 0.9378 0.9532 0.9876 0.9958 0.9892 0.9807 0.9915 0.9665<br />
P 11.98 13.90 7.526 4.801 5.425 8.631 5.840 13.05<br />
Es 2.479 2.290 1.644 0.8024 1.339 2.165 1.273 2.997<br />
Smith b -4.900 -5.816 -6.815 -7.494 -5.457 -6.517 -6.422 -6.187<br />
c 4.642 3.436 1.531 0.7796 3.430 2.119 1.918 1.674<br />
R 2 0.9432 0.9863 0.9972 0.9903 0.9965 0.9899 0.9926 0.9745<br />
P 13.14 11.06 2.799 9.100 3.434 4.745 4.769 6.834<br />
Es 1.996 1.135 0.6048 1.231 0.5348 1.088 0.9196 1.667<br />
Oswin b 8.662 8.177 6.951 6.559 7.783 7.375 6.943 6.821<br />
c 0.260 0.301 0.364 0.398 0.300 0.343 0.354 0.344<br />
R 2 0.936 0.978 0.992 0.999 0.987 0.978 0.992 0.955<br />
P 16.35 16.08 10.18 5.247 8.611 13.67 6.616 19.10<br />
Es 2.122 1.430 1.000 0.4352 1.029 1.611 0.9704 2.224<br />
Halsey b 2.816 2.412 2.121 2.057 2.584 2.196 2.260 2.090<br />
c 200.2 71.92 30.65 27.16 102.7 38.40 43.06 23.92<br />
R 2 0.9046 0.9119 0.9613 0.9785 0.9653 0.9534 0.9688 0.9314<br />
P 16.42 19.08 13.25 10.54 10.37 13.99 10.90 18.35<br />
Es 2.997 2.944 2.462 1.758 2.026 2.914 1.963 3.626<br />
Caurie II b 1.067 0.7904 0.5304 0.5116 0.9534 0.6279 0.6674 0.3633<br />
c 2.043 2.423 2.703 2.751 2.154 2.594 2.509 2.824<br />
R 2 0.9733 0.9811 0.9695 0.9492 0.9718 0.9749 0.9556 0.9822<br />
P 9.357 8.451 12.60 15.83 10.31 10.64 14.20 8.790<br />
Es 1.779 1.951 3.169 4.391 2.120 2.725 3.387 2.507<br />
Modified GAB b 5.705 4.784 3.440 3.007 4.080 3.864 3.282 3.798<br />
c 0.771 0.832 0.897 0.920 0.850 0.877 0.897 0.872<br />
d 322.894 503.164 1296.622 200000000 1364.002 694.361 1047.207 443.567<br />
R 2 0.960 0.985 0.995 0.992 0.998 0.990 0.992 0.980<br />
P 10.3181 7.6235 5.9209 7.7407 2.7014 5.071 8.349 5.437<br />
Es 1.8652 1.3230 0.8616 1.2204 0.4367 1.215 1.203 1.650<br />
Kuhn b -0.3184 -0.3773 -0.4441 -0.4953 -0.3627 -0.4264 -0.4204 -0.3849<br />
c 8.700 8.312 7.140 6.813 7.826 7.495 7.174 7.039<br />
R 2 0.7719 0.8408 0.8986 0.9396 0.8533 0.8587 0.9027 0.8194<br />
P 33.75 41.20 37.63 32.25 31.00 40.19 32.20 47.78<br />
Es 4.000 3.869 3.611 3.080 3.475 4.077 3.341 4.434<br />
Iglesias - Chirife b 0.319 0.378 0.444 0.496 0.363 0.427 0.421 0.385<br />
c 8.842 8.481 7.339 7.033 7.987 7.685 7.361 7.212<br />
R 2 0.770 0.840 0.898 0.939 0.852 0.858 0.902 0.818<br />
P 33.89 41.39 37.85 32.49 31.15 40.40 32.41 48.00<br />
Es 4.012 3.885 3.627 3.099 3.489 4.092 3.357 4.447<br />
Mizrahi b -8.842 -8.481 -7.339 -7.033 -7.987 -7.685 -7.361 -7.212<br />
c -8.523 -8.103 -6.894 -6.538 -7.624 -7.258 -6.941 -6.827<br />
R 2 0.770 0.840 0.898 0.939 0.852 0.858 0.902 0.818<br />
P 33.89 41.39 37.85 32.50 31.15 40.42 32.41 48.00<br />
Es 4.012 3.885 3.627 3.099 3.489 4.093 3.357 4.447<br />
Modified BET mo 2.164 2.336 2.453 2.608 2.234 2.433 2.366 2.220<br />
Note: m o, b, c and d are constants.<br />
b 30000000 70000000 50000000 70000000 60000000 40000000 50000000 40000000<br />
R 2 0.354 0.607 0.819 0.893 0.619 0.749 0.799 0.718<br />
P 43.43 36.75 30.12 26.05 39.39 33.36 33.33 33.08<br />
Es 6.730 6.082 4.830 4.091 5.597 5.429 4.799 5.535<br />
212