Colletotrichum: complex species or species ... - CBS - KNAW
Colletotrichum: complex species or species ... - CBS - KNAW
Colletotrichum: complex species or species ... - CBS - KNAW
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
The <strong>Colletotrichum</strong> gloeosp<strong>or</strong>ioides <strong>species</strong> <strong>complex</strong><br />
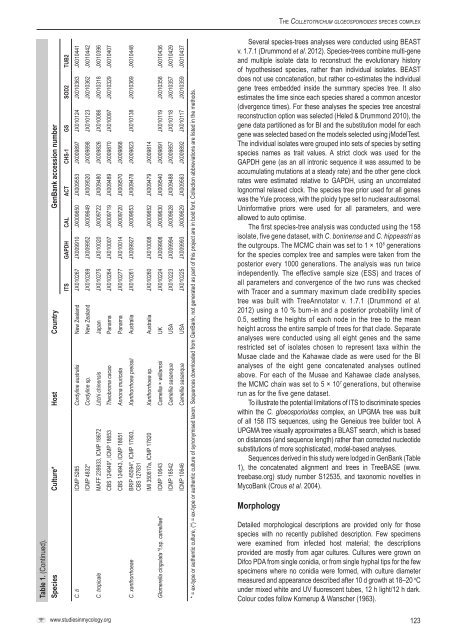
Table 1. (Continued).<br />
Species Culture* Host Country GenBank accession number<br />
ITS GAPDH CAL ACT CHS-1 GS SOD2 TUB2<br />
C. ti ICMP 5285 C<strong>or</strong>dyline australis New Zealand JX010267 JX009910 JX009650 JX009553 JX009897 JX010124 JX010363 JX010441<br />
ICMP 4832* C<strong>or</strong>dyline sp. New Zealand JX010269 JX009952 JX009649 JX009520 JX009898 JX010123 JX010362 JX010442<br />
C. tropicale MAFF 239933, ICMP 18672 Litchi chinensis Japan JX010275 JX010020 JX009722 JX009480 JX009826 JX010086 JX010318 JX010396<br />
<strong>CBS</strong> 124949*, ICMP 18653 Theobroma cacao Panama JX010264 JX010007 JX009719 JX009489 JX009870 JX010097 JX010329 JX010407<br />
<strong>CBS</strong> 124943, ICMP 18651 Annona muricata Panama JX010277 JX010014 JX009720 JX009570 JX009868<br />
Xanth<strong>or</strong>rhoea preissii Australia JX010261 JX009927 JX009653 JX009478 JX009823 JX010138 JX010369 JX010448<br />
C. xanth<strong>or</strong>rhoeae BRIP 45094*, ICMP 17903,<br />
<strong>CBS</strong> 127831<br />
IMI 350817a, ICMP 17820 Xanth<strong>or</strong>rhoea sp. Australia JX010260 JX010008 JX009652 JX009479 JX009814<br />
Glomerella cingulata “f.sp. camelliae” ICMP 10643 Camellia × williamsii UK JX010224 JX009908 JX009630 JX009540 JX009891 JX010119 JX010358 JX010436<br />
ICMP 18542 Camellia sasanqua USA JX010223 JX009994 JX009628 JX009488 JX009857 JX010118 JX010357 JX010429<br />
ICMP 10646 Camellia sasanqua USA JX010225 JX009993 JX009629 JX009563 JX009892 JX010117 JX010359 JX010437<br />
* = ex-type <strong>or</strong> authentic culture, (*) = ex-type <strong>or</strong> authentic culture of synonymised taxon. Sequences downloaded from GenBank, not generated as part of this project are in bold font. Collection abbreviations are listed in the methods.<br />
Several <strong>species</strong>-trees analyses were conducted using BEAST<br />
v. 1.7.1 (Drummond et al. 2012). Species-trees combine multi-gene<br />
and multiple isolate data to reconstruct the evolutionary hist<strong>or</strong>y<br />
of hypothesised <strong>species</strong>, rather than individual isolates. BEAST<br />
does not use concatenation, but rather co-estimates the individual<br />
gene trees embedded inside the summary <strong>species</strong> tree. It also<br />
estimates the time since each <strong>species</strong> shared a common ancest<strong>or</strong><br />
(divergence times). F<strong>or</strong> these analyses the <strong>species</strong> tree ancestral<br />
reconstruction option was selected (Heled & Drummond 2010), the<br />
gene data partitioned as f<strong>or</strong> BI and the substitution model f<strong>or</strong> each<br />
gene was selected based on the models selected using jModelTest.<br />
The individual isolates were grouped into sets of <strong>species</strong> by setting<br />
<strong>species</strong> names as trait values. A strict clock was used f<strong>or</strong> the<br />
GAPDH gene (as an all intronic sequence it was assumed to be<br />
accumulating mutations at a steady rate) and the other gene clock<br />
rates were estimated relative to GAPDH, using an unc<strong>or</strong>related<br />
logn<strong>or</strong>mal relaxed clock. The <strong>species</strong> tree pri<strong>or</strong> used f<strong>or</strong> all genes<br />
was the Yule process, with the ploidy type set to nuclear autosomal.<br />
Uninf<strong>or</strong>mative pri<strong>or</strong>s were used f<strong>or</strong> all parameters, and were<br />
allowed to auto optimise.<br />
The first <strong>species</strong>-tree analysis was conducted using the 158<br />
isolate, five gene dataset, with C. boninense and C. hippeastri as<br />
the outgroups. The MCMC chain was set to 1 × 10 8 generations<br />
f<strong>or</strong> the <strong>species</strong> <strong>complex</strong> tree and samples were taken from the<br />
posteri<strong>or</strong> every 1000 generations. The analysis was run twice<br />
independently. The effective sample size (ESS) and traces of<br />
all parameters and convergence of the two runs was checked<br />
with Tracer and a summary maximum clade credibility <strong>species</strong><br />
tree was built with TreeAnnotat<strong>or</strong> v. 1.7.1 (Drummond et al.<br />
2012) using a 10 % burn-in and a posteri<strong>or</strong> probability limit of<br />
0.5, setting the heights of each node in the tree to the mean<br />
height across the entire sample of trees f<strong>or</strong> that clade. Separate<br />
analyses were conducted using all eight genes and the same<br />
restricted set of isolates chosen to represent taxa within the<br />
Musae clade and the Kahawae clade as were used f<strong>or</strong> the BI<br />
analyses of the eight gene concatenated analyses outlined<br />
above. F<strong>or</strong> each of the Musae and Kahawae clade analyses,<br />
the MCMC chain was set to 5 × 10 7 generations, but otherwise<br />
run as f<strong>or</strong> the five gene dataset.<br />
To illustrate the potential limitations of ITS to discriminate <strong>species</strong><br />
within the C. gloeosp<strong>or</strong>ioides <strong>complex</strong>, an UPGMA tree was built<br />
of all 158 ITS sequences, using the Geneious tree builder tool. A<br />
UPGMA tree visually approximates a BLAST search, which is based<br />
on distances (and sequence length) rather than c<strong>or</strong>rected nucleotide<br />
substitutions of m<strong>or</strong>e sophisticated, model-based analyses.<br />
Sequences derived in this study were lodged in GenBank (Table<br />
1), the concatenated alignment and trees in TreeBASE (www.<br />
treebase.<strong>or</strong>g) study number S12535, and taxonomic novelties in<br />
MycoBank (Crous et al. 2004).<br />
M<strong>or</strong>phology<br />
Detailed m<strong>or</strong>phological descriptions are provided only f<strong>or</strong> those<br />
<strong>species</strong> with no recently published description. Few specimens<br />
were examined from infected host material; the descriptions<br />
provided are mostly from agar cultures. Cultures were grown on<br />
Difco PDA from single conidia, <strong>or</strong> from single hyphal tips f<strong>or</strong> the few<br />
specimens where no conidia were f<strong>or</strong>med, with culture diameter<br />
measured and appearance described after 10 d growth at 18–20 o C<br />
under mixed white and UV flu<strong>or</strong>escent tubes, 12 h light/12 h dark.<br />
Colour codes follow K<strong>or</strong>nerup & Wanscher (1963).<br />
www.studiesinmycology.<strong>or</strong>g<br />
123