Colletotrichum: complex species or species ... - CBS - KNAW
Colletotrichum: complex species or species ... - CBS - KNAW
Colletotrichum: complex species or species ... - CBS - KNAW
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
The <strong>Colletotrichum</strong> gloeosp<strong>or</strong>ioides <strong>species</strong> <strong>complex</strong><br />
A<br />
1<br />
1<br />
1<br />
0.95<br />
0.97<br />
1<br />
0.99<br />
ICMP 18613 Limonium Israel<br />
ICMP 18727 Fragaria USA<br />
ICMP 18581 C. fructicola Coffea Thailand<br />
ICMP 18646 C. ignotum Tetragastris Panama<br />
ICMP 17921 Glomerella cingulata var. min<strong>or</strong> Ficus Germany<br />
ICMP 18645<br />
Theobroma Panama<br />
ICMP 18120 Diosc<strong>or</strong>ea Nigeria<br />
C. fructicola<br />
1<br />
1<br />
0.69<br />
ICMP 17938 Nuphar USA<br />
ICMP 18187 C. nupharicola Nuphar USA<br />
ICMP 17940 Nuphar USA<br />
C. nupharicola<br />
0.99<br />
1<br />
1<br />
1<br />
1<br />
ICMP 18621 Persea New Zealand<br />
ICMP 12071 C. alienum Malus New Zealand<br />
ICMP 18686 Pyrus Japan<br />
ICMP 18691 Persea Australia<br />
ICMP 18608 C. aenigma Persea Israel<br />
C. alienum<br />
C. aenigma<br />
1<br />
ICMP 17817 Musa Kenya<br />
ICMP 19119 C. musae Musa USA<br />
C. musae<br />
0.94<br />
1<br />
0.82<br />
1<br />
1<br />
1<br />
ICMP 17795 Malus USA<br />
ICMP 18642 C. hymenocallidis Hymenocallis China<br />
ICMP 18578 C. siamense Coffee Thailand<br />
ICMP 19118 C. jasmini-sambac Jasminum Vietnam<br />
ICMP 18574 Pistacia Australia<br />
C. siamense<br />
1<br />
0.87<br />
1<br />
1<br />
1<br />
1<br />
1<br />
1<br />
1<br />
ICMP 12567 Persea Australia<br />
ICMP 18121 Diosc<strong>or</strong>ia Nigeria<br />
ICMP 17673 C.g. “f. sp. aeschynomenes”Aeschynomene USA<br />
ICMP 18672 Litchi Japan<br />
ICMP 18653 C. tropicale Theobroma Panama<br />
ICMP 18705 Coffea Fiji<br />
ICMP 1778 C. gloeosp<strong>or</strong>ioides var. minus Carica Australia<br />
ICMP 19051 C.g. “f. sp. salsolae” Salsola Hungary<br />
ICMP 18696 Mangifera Australia<br />
ICMP 18580 C. asianum Coffea Thailand<br />
ICMP 17821 C. gloeosp<strong>or</strong>ioides Citrus Italy<br />
C. aeschynomenes<br />
C. tropicale<br />
C. queenslandicum<br />
C. salsolae<br />
C. asianum<br />
0.0030<br />
B<br />
1<br />
1<br />
0.44<br />
0.55<br />
0.33<br />
0.47<br />
0.75<br />
0.90<br />
0.97<br />
0.97<br />
0.60<br />
C. fructicola<br />
C. alienum<br />
C. nupharicola<br />
C. aenigma<br />
C. musae<br />
C. aeschynomenes<br />
C. siamense<br />
C. tropicale<br />
C. salsolae<br />
C. queenslandicum<br />
C. asianum<br />
C. gloeosp<strong>or</strong>ioides<br />
0.13<br />
0.117<br />
0.104<br />
0.091<br />
0.078<br />
0.065<br />
0.052<br />
0.039<br />
0.026<br />
0.013<br />
0.0<br />
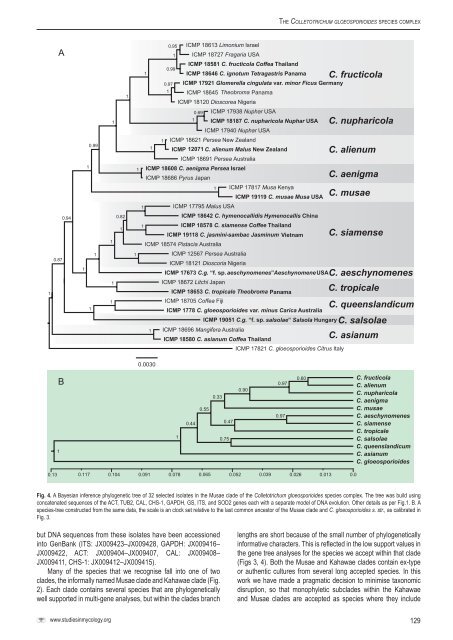
Fig. 4. A Bayesian inference phylogenetic tree of 32 selected isolates in the Musae clade of the <strong>Colletotrichum</strong> gloeosp<strong>or</strong>ioides <strong>species</strong> <strong>complex</strong>. The tree was build using<br />
concatenated sequences of the ACT, TUB2, CAL, CHS-1, GAPDH, GS, ITS, and SOD2 genes each with a separate model of DNA evolution. Other details as per Fig.1. B. A<br />
<strong>species</strong>-tree constructed from the same data, the scale is an clock set relative to the last common ancest<strong>or</strong> of the Musae clade and C. gloeosp<strong>or</strong>ioides s. str., as calibrated in<br />
Fig. 3.<br />
but DNA sequences from these isolates have been accessioned<br />
into GenBank (ITS: JX009423–JX009428, GAPDH: JX009416–<br />
JX009422, ACT: JX009404–JX009407, CAL: JX009408–<br />
JX009411, CHS-1: JX009412–JX009415).<br />
Many of the <strong>species</strong> that we recognise fall into one of two<br />
clades, the inf<strong>or</strong>mally named Musae clade and Kahawae clade (Fig.<br />
2). Each clade contains several <strong>species</strong> that are phylogenetically<br />
well supp<strong>or</strong>ted in multi-gene analyses, but within the clades branch<br />
lengths are sh<strong>or</strong>t because of the small number of phylogenetically<br />
inf<strong>or</strong>mative characters. This is reflected in the low supp<strong>or</strong>t values in<br />
the gene tree analyses f<strong>or</strong> the <strong>species</strong> we accept within that clade<br />
(Figs 3, 4). Both the Musae and Kahawae clades contain ex-type<br />
<strong>or</strong> authentic cultures from several long accepted <strong>species</strong>. In this<br />
w<strong>or</strong>k we have made a pragmatic decision to minimise taxonomic<br />
disruption, so that monophyletic subclades within the Kahawae<br />
and Musae clades are accepted as <strong>species</strong> where they include<br />
www.studiesinmycology.<strong>or</strong>g<br />
129