Colletotrichum: complex species or species ... - CBS - KNAW
Colletotrichum: complex species or species ... - CBS - KNAW
Colletotrichum: complex species or species ... - CBS - KNAW
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Damm et al.<br />
1 change<br />
99<br />
1.00<br />
86<br />
1.00<br />
0.96<br />
98<br />
1.00<br />
0.98<br />
0.98<br />
93<br />
1.00<br />
72<br />
98<br />
1.00<br />
79<br />
1.00<br />
98<br />
1.00<br />
<strong>CBS</strong> 515.78 Nymphaea NL<br />
<strong>CBS</strong> 516.78 Nuphar NL<br />
<strong>CBS</strong> 526.77 Nymphaea NL<br />
<strong>CBS</strong> 173.51 Mahonia Italy<br />
<strong>CBS</strong> 129945 Olea P<strong>or</strong>tugal<br />
<strong>CBS</strong> 231.49 Olea P<strong>or</strong>tugal<br />
<strong>CBS</strong> 126507 Oenothera NL<br />
IMI 370491 Malus Brazil<br />
<strong>CBS</strong> 119294 Leucaena Mexico<br />
<strong>CBS</strong> 112992 Protea ZA<br />
<strong>CBS</strong> 113002 Protea ZA<br />
<strong>CBS</strong> 113003 Protea ZA<br />
<strong>CBS</strong> 113004 Protea ZA<br />
<strong>CBS</strong> 113005 Protea ZA<br />
<strong>CBS</strong> 115408 Protea ZA<br />
<strong>CBS</strong> 482.82 Protea Australia<br />
<strong>CBS</strong> 114188 Protea unknown<br />
<strong>CBS</strong> 158.27 unknown unknown<br />
CSL 455 Photinia UK<br />
IMI 379162 Capsicum Zimbabwe<br />
IMI 360386 Pelargonium India<br />
<strong>CBS</strong> 129926 litter Thailand<br />
<strong>CBS</strong> 126528 Capsicum Indonesia<br />
<strong>CBS</strong> 126382 Anemone NL<br />
<strong>CBS</strong> 126383 Anemone NL<br />
<strong>CBS</strong> 126511 Anemone NL<br />
<strong>CBS</strong> 100064 Anemone NL<br />
<strong>CBS</strong> 130.80 Anemone Italy<br />
<strong>CBS</strong> 129935 Anemone Israel<br />
<strong>CBS</strong> 127612 Fragaria USA<br />
<strong>CBS</strong> 361.79 Anemone NL<br />
<strong>CBS</strong> 126513 Anemone NL<br />
<strong>CBS</strong> 100065 Anemone NL<br />
<strong>CBS</strong> 129928 Fragaria USA<br />
<strong>CBS</strong> 129929 Fragaria USA<br />
<strong>CBS</strong> 112202 Fragaria Spain<br />
<strong>CBS</strong> 122110 Fragaria Bulgaria<br />
<strong>CBS</strong> 122111 Fragaria Bulgaria<br />
<strong>CBS</strong> 122121 Fragaria Bulgaria<br />
IMI 345053 Fragaria France<br />
IMI 348497 Fragaria France<br />
<strong>CBS</strong> 126372 Fragaria Italy<br />
IMI 360928 Fragaria Switzerland<br />
IMI 364856 Fragaria Spain<br />
<strong>CBS</strong> 125958 Fragaria NL<br />
<strong>CBS</strong> 125959 Fragaria NL<br />
<strong>CBS</strong> 125966 Fragaria NL<br />
<strong>CBS</strong> 125961 Fragaria NL<br />
<strong>CBS</strong> 130239 Fragaria NL<br />
IMI 391664 Fragaria Israel<br />
<strong>CBS</strong> 129936 Fragaria Israel<br />
<strong>CBS</strong> 129937 Fragaria Israel<br />
<strong>CBS</strong> 129918 Fragaria unknown<br />
<strong>CBS</strong> 126366 Fragaria USA<br />
<strong>CBS</strong> 126367 Fragaria USA<br />
<strong>CBS</strong> 126370 Fragaria USA<br />
IMI 311743 Fragaria USA<br />
IMI 324995 Fragaria USA<br />
<strong>CBS</strong> 127609 Fragaria USA<br />
<strong>CBS</strong> 127610 Fragaria USA<br />
<strong>CBS</strong> 129933 Fragaria USA<br />
IMI 348177 Fragaria USA<br />
<strong>CBS</strong> 127608 Fragaria Canada<br />
<strong>CBS</strong> 126504 Fragaria ZA<br />
IMI 301119 Fragaria Kenya<br />
IMI 299103 Fragaria UK<br />
<strong>CBS</strong> 125973 Fragaria UK<br />
<strong>CBS</strong> 126377 Fragaria NL<br />
IMI 348502 Fragaria France<br />
IMI 345032 Fragaria Italy<br />
99 <strong>CBS</strong> 126529 Capsicum Indonesia<br />
98 1.00<br />
<strong>CBS</strong> 126530 Capsicum Indonesia<br />
C. scovillei<br />
<strong>CBS</strong> 120708 Capsicum Thailand<br />
1.00 IMI 350839 Psidium India<br />
C. guajavae<br />
100 <strong>CBS</strong> 126518 Carthamus NL<br />
<strong>CBS</strong> 126519 Chrysanthemum NL C. chrysanthemi<br />
1.00 IMI 364540 Chrysanthemum China<br />
90 <strong>CBS</strong> 853.73 Cosmos NL<br />
C. cosmi<br />
1.00 <strong>CBS</strong> 125472 Coffea Vietnam<br />
C. walleri<br />
<strong>CBS</strong> 122122 Carica Australia<br />
0.99 <strong>CBS</strong> 294.67 Carica Australia<br />
<strong>CBS</strong> 295.67 Fragaria Australia<br />
IMI 345034 Fragaria Australia<br />
IMI 354381 Fragaria Australia C. simmondsii<br />
99 <strong>CBS</strong> 126524 Cyclamen NL<br />
1.00<br />
<strong>CBS</strong> 111531 Protea USA<br />
99 1.00 97<br />
1.00 <strong>CBS</strong> 114494 Protea USA<br />
IMI 313840 Mangifera Australia<br />
97 1.00<br />
99 IMI 165753 Musa St. Lucia<br />
1.00 1.00 <strong>CBS</strong> 502.97 Musa West Indies<br />
IMI 364297 Theobroma Malaysia<br />
<strong>CBS</strong> 127551 Eucalyptus Indonesia<br />
99 <strong>CBS</strong> 112989 Hevea India<br />
1.00 <strong>CBS</strong> 129827 Hevea Colombia<br />
<strong>CBS</strong> 292.67 Capsicum Australia<br />
C. nymphaeae<br />
C. paxtonii<br />
C. sloanei<br />
C. indonesiense<br />
C. laticiphilum<br />
C. brisbanense<br />
A2<br />
D2<br />
D4<br />
D3<br />
Clade 2<br />
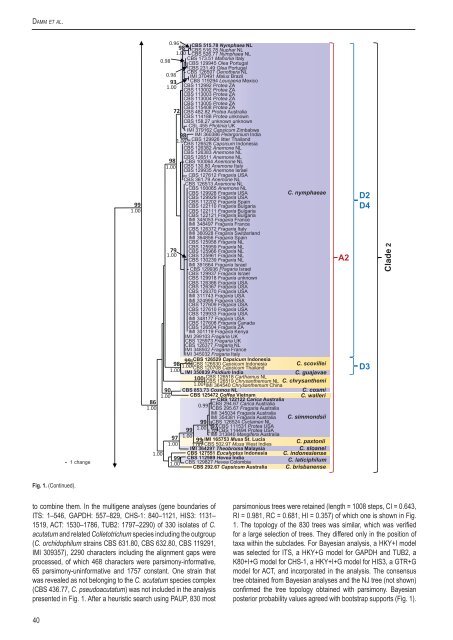
Fig. 1. (Continued).<br />
to combine them. In the multigene analyses (gene boundaries of<br />
ITS: 1–546, GAPDH: 557–829, CHS-1: 840–1121, HIS3: 1131–<br />
1519, ACT: 1530–1786, TUB2: 1797–2290) of 330 isolates of C.<br />
acutatum and related <strong>Colletotrichum</strong> <strong>species</strong> including the outgroup<br />
(C. <strong>or</strong>chidophilum strains <strong>CBS</strong> 631.80, <strong>CBS</strong> 632.80, <strong>CBS</strong> 119291,<br />
IMI 309357), 2290 characters including the alignment gaps were<br />
processed, of which 468 characters were parsimony-inf<strong>or</strong>mative,<br />
65 parsimony-uninf<strong>or</strong>mative and 1757 constant. One strain that<br />
was revealed as not belonging to the C. acutatum <strong>species</strong> <strong>complex</strong><br />
(<strong>CBS</strong> 436.77, C. pseudoacutatum) was not included in the analysis<br />
presented in Fig. 1. After a heuristic search using PAUP, 830 most<br />
parsimonious trees were retained (length = 1008 steps, CI = 0.643,<br />
RI = 0.981, RC = 0.681, HI = 0.357) of which one is shown in Fig.<br />
1. The topology of the 830 trees was similar, which was verified<br />
f<strong>or</strong> a large selection of trees. They differed only in the position of<br />
taxa within the subclades. F<strong>or</strong> Bayesian analysis, a HKY+I model<br />
was selected f<strong>or</strong> ITS, a HKY+G model f<strong>or</strong> GAPDH and TUB2, a<br />
K80+I+G model f<strong>or</strong> CHS-1, a HKY+I+G model f<strong>or</strong> HIS3, a GTR+G<br />
model f<strong>or</strong> ACT, and inc<strong>or</strong>p<strong>or</strong>ated in the analysis. The consensus<br />
tree obtained from Bayesian analyses and the NJ tree (not shown)<br />
confirmed the tree topology obtained with parsimony. Bayesian<br />
posteri<strong>or</strong> probability values agreed with bootstrap supp<strong>or</strong>ts (Fig. 1).<br />
40