Colletotrichum: complex species or species ... - CBS - KNAW
Colletotrichum: complex species or species ... - CBS - KNAW
Colletotrichum: complex species or species ... - CBS - KNAW
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
Weir et al.<br />
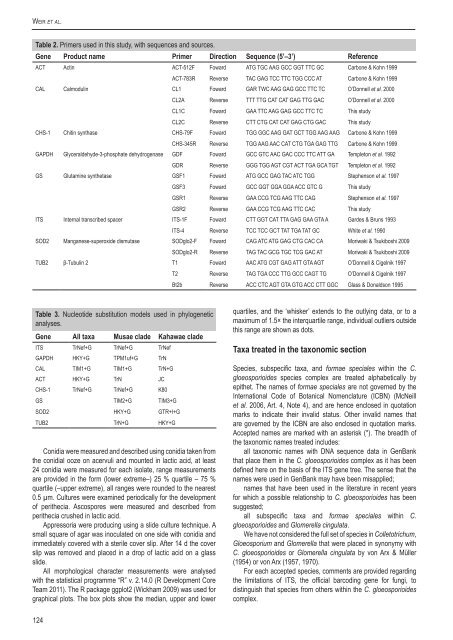
Table 2. Primers used in this study, with sequences and sources.<br />
Gene Product name Primer Direction Sequence (5’–3’) Reference<br />
ACT Actin ACT-512F Foward ATG TGC AAG GCC GGT TTC GC Carbone & Kohn 1999<br />
ACT-783R Reverse TAC GAG TCC TTC TGG CCC AT Carbone & Kohn 1999<br />
CAL Calmodulin CL1 Foward GAR TWC AAG GAG GCC TTC TC O’Donnell et al. 2000<br />
CL2A Reverse TTT TTG CAT CAT GAG TTG GAC O’Donnell et al. 2000<br />
CL1C Foward GAA TTC AAG GAG GCC TTC TC This study<br />
CL2C Reverse CTT CTG CAT CAT GAG CTG GAC This study<br />
CHS-1 Chitin synthase CHS-79F Foward TGG GGC AAG GAT GCT TGG AAG AAG Carbone & Kohn 1999<br />
CHS-345R Reverse TGG AAG AAC CAT CTG TGA GAG TTG Carbone & Kohn 1999<br />
GAPDH Glyceraldehyde-3-phosphate dehydrogenase GDF Foward GCC GTC AAC GAC CCC TTC ATT GA Templeton et al. 1992<br />
GDR Reverse GGG TGG AGT CGT ACT TGA GCA TGT Templeton et al. 1992<br />
GS Glutamine synthetase GSF1 Foward ATG GCC GAG TAC ATC TGG Stephenson et al. 1997<br />
GSF3 Foward GCC GGT GGA GGA ACC GTC G This study<br />
GSR1 Reverse GAA CCG TCG AAG TTC CAG Stephenson et al. 1997<br />
GSR2 Reverse GAA CCG TCG AAG TTC CAC This study<br />
ITS Internal transcribed spacer ITS-1F Foward CTT GGT CAT TTA GAG GAA GTA A Gardes & Bruns 1993<br />
ITS-4 Reverse TCC TCC GCT TAT TGA TAT GC White et al. 1990<br />
SOD2 Manganese-superoxide dismutase SODglo2-F Foward CAG ATC ATG GAG CTG CAC CA M<strong>or</strong>iwaki & Tsukiboshi 2009<br />
SODglo2-R Reverse TAG TAC GCG TGC TCG GAC AT M<strong>or</strong>iwaki & Tsukiboshi 2009<br />
TUB2 β-Tubulin 2 T1 Foward AAC ATG CGT GAG ATT GTA AGT O’Donnell & Cigelnik 1997<br />
T2 Reverse TAG TGA CCC TTG GCC CAGT TG O’Donnell & Cigelnik 1997<br />
Bt2b Reverse ACC CTC AGT GTA GTG ACC CTT GGC Glass & Donaldson 1995<br />
Table 3. Nucleotide substitution models used in phylogenetic<br />
analyses.<br />
Gene All taxa Musae clade Kahawae clade<br />
ITS TrNef+G TrNef+G TrNef<br />
GAPDH HKY+G TPM1uf+G TrN<br />
CAL TIM1+G TIM1+G TrN+G<br />
ACT HKY+G TrN JC<br />
CHS-1 TrNef+G TrNef+G K80<br />
GS TIM2+G TIM3+G<br />
SOD2 HKY+G GTR+I+G<br />
TUB2 TrN+G HKY+G<br />
Conidia were measured and described using conidia taken from<br />
the conidial ooze on acervuli and mounted in lactic acid, at least<br />
24 conidia were measured f<strong>or</strong> each isolate, range measurements<br />
are provided in the f<strong>or</strong>m (lower extreme–) 25 % quartile – 75 %<br />
quartile (–upper extreme), all ranges were rounded to the nearest<br />
0.5 µm. Cultures were examined periodically f<strong>or</strong> the development<br />
of perithecia. Ascosp<strong>or</strong>es were measured and described from<br />
perithecia crushed in lactic acid.<br />
Appress<strong>or</strong>ia were producing using a slide culture technique. A<br />
small square of agar was inoculated on one side with conidia and<br />
immediately covered with a sterile cover slip. After 14 d the cover<br />
slip was removed and placed in a drop of lactic acid on a glass<br />
slide.<br />
All m<strong>or</strong>phological character measurements were analysed<br />
with the statistical programme “R” v. 2.14.0 (R Development C<strong>or</strong>e<br />
Team 2011). The R package ggplot2 (Wickham 2009) was used f<strong>or</strong><br />
graphical plots. The box plots show the median, upper and lower<br />
quartiles, and the ‘whisker’ extends to the outlying data, <strong>or</strong> to a<br />
maximum of 1.5× the interquartile range, individual outliers outside<br />
this range are shown as dots.<br />
Taxa treated in the taxonomic section<br />
Species, subspecific taxa, and f<strong>or</strong>mae speciales within the C.<br />
gloeosp<strong>or</strong>ioides <strong>species</strong> <strong>complex</strong> are treated alphabetically by<br />
epithet. The names of f<strong>or</strong>mae speciales are not governed by the<br />
International Code of Botanical Nomenclature (ICBN) (McNeill<br />
et al. 2006, Art. 4, Note 4), and are hence enclosed in quotation<br />
marks to indicate their invalid status. Other invalid names that<br />
are governed by the ICBN are also enclosed in quotation marks.<br />
Accepted names are marked with an asterisk (*). The breadth of<br />
the taxonomic names treated includes:<br />
all taxonomic names with DNA sequence data in GenBank<br />
that place them in the C. gloeosp<strong>or</strong>ioides <strong>complex</strong> as it has been<br />
defined here on the basis of the ITS gene tree. The sense that the<br />
names were used in GenBank may have been misapplied;<br />
names that have been used in the literature in recent years<br />
f<strong>or</strong> which a possible relationship to C. gloeosp<strong>or</strong>ioides has been<br />
suggested;<br />
all subspecific taxa and f<strong>or</strong>mae speciales within C.<br />
gloeosp<strong>or</strong>ioides and Glomerella cingulata.<br />
We have not considered the full set of <strong>species</strong> in <strong>Colletotrichum</strong>,<br />
Gloeosp<strong>or</strong>ium and Glomerella that were placed in synonymy with<br />
C. gloeosp<strong>or</strong>ioides <strong>or</strong> Glomerella cingulata by von Arx & Müller<br />
(1954) <strong>or</strong> von Arx (1957, 1970).<br />
F<strong>or</strong> each accepted <strong>species</strong>, comments are provided regarding<br />
the limitations of ITS, the official barcoding gene f<strong>or</strong> fungi, to<br />
distinguish that <strong>species</strong> from others within the C. gloeosp<strong>or</strong>ioides<br />
<strong>complex</strong>.<br />
124