Drosophila - Severo Ochoa - Universidad Autónoma de Madrid
Drosophila - Severo Ochoa - Universidad Autónoma de Madrid
Drosophila - Severo Ochoa - Universidad Autónoma de Madrid
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Jefe <strong>de</strong> Línea /<br />
Group Lea<strong>de</strong>r:<br />
José Manuel Sierra<br />
Personal Científico /<br />
Scientific Staff:<br />
José María Izquierdo<br />
Becarios Predoctorales /<br />
Predoctoral Fellows:<br />
Raquel Reyes Manzanas<br />
Técnicos <strong>de</strong> Investigación /<br />
Technical Assistance:<br />
José Alcal<strong>de</strong><br />
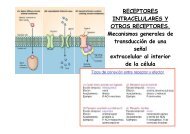
Regulación <strong>de</strong> la expresión génica Regulation of gene expression<br />
Bases moleculares <strong>de</strong> la regulación<br />
post-transcripcional en eucariotas<br />
Resumen <strong>de</strong> investigación<br />
Nuestro grupo <strong>de</strong> investigación está interesado en el<br />
estudio <strong>de</strong> las bases moleculares <strong>de</strong> la regulación<br />
post-transcripcional en eucariotas y, en concreto, en los<br />
mecanismos <strong>de</strong>l "splicing" <strong>de</strong> intrones y <strong>de</strong> la traducción <strong>de</strong><br />
RNA mensajeros. Deseamos conocer prioritariamente el<br />
papel que estos procesos juegan en el origen y progresión<br />
<strong>de</strong>l cáncer, así como en la diferenciación celular durante el<br />
<strong>de</strong>sarrollo embrionario.<br />
Factores proteicos implicados en el reconocimiento<br />
<strong>de</strong>l mRNA y su papel en el <strong>de</strong>sarrollo embrionario y<br />
proliferación celular.<br />
Las proteínas eIF (eukaryotic initiation factor) 4E, eIF4G,<br />
eIF4A y eIF4B están implicadas en el reconocimiento y<br />
unión <strong>de</strong>l mRNA al ribosoma. Por lo que se refiere al eIF4E,<br />
resultados previos <strong>de</strong> nuestro laboratorio <strong>de</strong>mostraron que<br />
en <strong>Drosophila</strong> existen siete genes que codifican ocho<br />
isoformas distintas <strong>de</strong> esta proteína. Uno <strong>de</strong> estos genes,<br />
eIF4E1/2, origina por “splicing” alternativo tres mRNAs y<br />
dos isoformas proteicas, eIF4E1 y eIF4E2, que difieren en<br />
sus extremos amino. Es interesante que en embriones<br />
carentes <strong>de</strong>l gen eIF4E1/2 se produce la inducción <strong>de</strong><br />
apoptosis y la traducción <strong>de</strong> varios mRNAs por un<br />
mecanismo in<strong>de</strong>pendiente <strong>de</strong>l reconocimiento <strong>de</strong> la<br />
estructura m 7 GpppN (“cap”). Nuestro trabajo mas reciente<br />
se ha centrado en el estudio <strong>de</strong> las bases moleculares <strong>de</strong>l<br />
“splicing” alternativo <strong>de</strong>l eIF4E1/2 <strong>de</strong> <strong>Drosophila</strong> y su<br />
posible relevancia biológica durante el <strong>de</strong>sarrollo.<br />
El papel <strong>de</strong> TIA-1 y TIAR en la regulación post-transcripcional<br />
<strong>de</strong> la oncogénesis.<br />
TIA-1 (“T-cell Internal Antigen-1”) y TIAR (“TIA-1-Related<br />
protein”) son dos reguladores <strong>de</strong> “splicing” alternativo, y <strong>de</strong><br />
otros procesos celulares a nivel post-transcripcional, cuyo<br />
potencial apoptótico previsiblemente se <strong>de</strong>sregula en<br />
cáncer. El objetivo <strong>de</strong> este proyecto <strong>de</strong> investigación es: I)<br />
i<strong>de</strong>ntificar RNAs y proteínas humanos que interaccionan<br />
con TIA-1 y TIAR; II) <strong>de</strong>terminar los mecanismos<br />
moleculares en los que están implicadas estas proteínas; III)<br />
conocer el comportamiento <strong>de</strong> estos factores durante el<br />
proceso <strong>de</strong> oncogénesis. La consecución <strong>de</strong> estos<br />
objetivos abre la posibilidad <strong>de</strong> <strong>de</strong>finir nuevas dianas que<br />
permitan intervenir para paliar la inci<strong>de</strong>ncia y/o progresión<br />
<strong>de</strong> enfermeda<strong>de</strong>s neoplásicas, inflamatorias o autoinmunes,<br />
así como <strong>de</strong>sór<strong>de</strong>nes neuro<strong>de</strong>generativos que, en<br />
humanos, son consecuencia <strong>de</strong> la <strong>de</strong>sregulación <strong>de</strong>l<br />
balance entre proliferación celular y apoptosis. Estos<br />
estudios, a<strong>de</strong>más, pue<strong>de</strong>n constituir uno <strong>de</strong> los primeros<br />
análisis <strong>de</strong>tallados <strong>de</strong>l papel que dos proteínas implicadas<br />
en apoptosis juegan en la regulación <strong>de</strong> la expresión génica<br />
a nivel post-transcripcional.<br />
Molecular bases of post-transcriptional<br />
regulation in eukaryotes<br />
Research summary<br />
Our research group is interested in the study of the<br />
molecular bases of the post-transcriptional regulation in<br />
eucaryotes and, in particular, in the mechanisms of intron<br />
splicing and mRNA translation. We wish to know primarily<br />
the role of these processes in the origin and progress of<br />
cancer, as well as in cell differentiation during embryonic<br />
<strong>de</strong>velopment.<br />
Protein factors involved in mRNA recognition and their role in<br />
embryonic <strong>de</strong>velopment and cell proliferation.<br />
The proteins eIF (eukaryotic initiation factor) 4E, eIF4G,<br />
eIF4A and eIF4B are involved in the recognition and binding<br />
of the mRNA to the ribosome. In relation to eIF4E, previous<br />
results from our laboratory showed that in <strong>Drosophila</strong> there<br />
are seven genes encoding eigth different isoforms of this<br />
protein. One of these genes, eIF4E1/2, gives rise by<br />
alternative splicing three mRNAs and two protein isoforms,<br />
eIF4E1 and eIF4E2, whose amino termini are different.<br />
Interestingly, in embryos lacking eIF4E1/2 gene, an<br />
induction of apoptosis as well as the translation of several<br />
mRNAs by a mechanism in<strong>de</strong>pen<strong>de</strong>nt of the recognition of<br />
the m 7 GpppN (cap) structure were observed. Our recent<br />
work has focused on the study of the molecular bases of the<br />
alternative splicing of <strong>Drosophila</strong> eIF4E1/2 and its possible<br />
biological relevance during <strong>de</strong>velopment.<br />
The role of TIA-1 and TIAR on the post-transcriptional<br />
regulation in oncogenesis.<br />
TIA-1 (“T-cell Internal Antigen-1”) y TIAR (“TIA-1-Related<br />
protein”) are two alternative splicing regulators, as well as<br />
other cellular processes at the post-transcriptional level,<br />
whose apoptotic features in cancer are properly altered. The<br />
goal of this research project is: I) to i<strong>de</strong>ntify human RNAs<br />
and proteins that interact with TIA-1 and TIAR; II) to study<br />
the molecular fine-tuning mechanisms where these proteins<br />
are implied; III) to un<strong>de</strong>rstand the behaviour of these factors<br />
in oncogenesis. Presumably, this project will open the<br />
possibility of <strong>de</strong>fining potential targets aimed to avoid the<br />
<strong>de</strong>velopment and/or progress of certain human pathologies<br />
such as cancer, inflammatory and autoimmune diseases as<br />
well as neuro<strong>de</strong>generative disor<strong>de</strong>rs, where a dysregulation<br />
by imbalanced cell proliferation and apoptosis has been<br />
<strong>de</strong>scribed. Furthermore, it will be one of the first <strong>de</strong>tailed<br />
analyses of regulation of programmed cell <strong>de</strong>ath at the posttranscriptional<br />
level.<br />
E15<br />
Publicaciones<br />
Publications<br />
Izquierdo, J.M., Majós, N., Bonnal, S., Martínez, C., Castelo, R.,Guigó,<br />
R., Bilbao, D. and Valcárcel, J. (2005). Regulation of Fas alternative<br />
splicing by antagonistic effects of TIA-1 and PTB on exon <strong>de</strong>finition.<br />
Mol. Cell. 19, 475-484.<br />
Roesler, J., Izquierdo, J.M., Ryser, M., Rosen-Wolff, A., Gahr, M.,<br />
Valcárcel, J., Lenardo, M.J. and Zheng, L. (2005). Haploinsufficiency,<br />
rather than the effect of an excessive production of soluble CD95<br />
(CD95{Delta}TM), is the basis for ALPS Ia in a family with duplicated 3'<br />
splice site AG in CD95 intron 5 on one allele. Blood. 106, 1652-1659.<br />
Hernán<strong>de</strong>z, G., Altmann, M., Sierra, J. M., Urlaub, H., Diez <strong>de</strong>l Corral,<br />
R., Schwartz, P. and Rivera-Pomar, R. (2005). Functional analysis of<br />
seven genes encoding eight translation initiation factor 4E (eIF4E)<br />
isoforms in <strong>Drosophila</strong>. Mech. of Develop. 122, 529-543.<br />
Izquierdo, J. M. and Cuezva, J. M. (2005). Epigenetic regulation<br />
of the binding activity of translation inhibitory proteins that bind the 3´<br />
untranslated region of beta-F1-ATPase mRNA by a<strong>de</strong>nine nucleoti<strong>de</strong>s<br />
and the redox state. Arch. Biochem. Biophys. 433, 481-486.<br />
Izquierdo, J.M. (2006). Control of the ATP synthase beta subunit<br />
expression by RNA-binding proteins TIA-1, TIAR, and HuR.<br />
Biochem. Biophys. Res. Commun. 348, 703-711.<br />
Izquierdo, J.M. and Valcárcel, J. (2006). A simple principle to explain<br />
the evolution of pre-mRNA splicing. Genes Dev. 20, 1679-1684.<br />
Figura 2. Sobreexpresión ectópica <strong>de</strong>l eIF4E1 en <strong>Drosophila</strong><br />
melanogaster.- Mosca con genotipo silvestre (A) o con el<br />
genotipo UAS-4E1/GAL4-NGT31;TM2/+ (B). Los halterios (H)<br />
silvestres o transformados en alas se indican con flechas.<br />
Figure 2. Ectopic overexpression of eIF4E1 in <strong>Drosophila</strong><br />
melanogaster.- Flies with wild-type (A) or UAS-4E1/GAL4-<br />
NGT31;TM2/+ (B) genotypes. Wild-type halteres (H) or<br />
transformed in wings are shown with arrows.<br />
Figura 1. Estructura <strong>de</strong>l gen eIF4E1/2 <strong>de</strong> <strong>Drosophila</strong> y esquema <strong>de</strong>l “splicing” alternativo.- Se indican las posiciones <strong>de</strong> las<br />
dianas EcoRI (R) en el fragmento <strong>de</strong> DNA. Los mRNA1 y mRNA2 codifican el eIF4E1, mientras que la isoforma eIF4E2 está<br />
codificada por el mRNA3. Los extremos amino específicos <strong>de</strong> cada isoforma están coloreados <strong>de</strong> azul y ver<strong>de</strong>.<br />
CBM 2005/2006<br />
142<br />
Figure 1. Structure of <strong>Drosophila</strong> eIF4E1/2 gene and outline of the alternative splicing.- The positions of restriction sites for<br />
EcoRI (R) are shown in the DNA fragment. The mRNA1 and mRNA2 enco<strong>de</strong> eIF4E1, while the eIF4E2 isoform is enco<strong>de</strong>d<br />
by mRNA3. The specific N-termini of each isoform are colored in blue and green.<br />
143