The Development of Novel Antibiotics Using ... - Jacobs University
The Development of Novel Antibiotics Using ... - Jacobs University
The Development of Novel Antibiotics Using ... - Jacobs University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
6<br />
<strong>The</strong> newly synthesized receptors showed a remarkable degree <strong>of</strong><br />
flexibility in comparison with macrocycles A or B. In addition<br />
they are chiral and can be obtained in almost quantitative yields<br />
as analytically and enantiomerically pure products. This study is<br />
a preliminary assessment <strong>of</strong> the recognition ability <strong>of</strong> the novel<br />
receptors to oligopeptides in the gas phase and further quantitative<br />
work aimed at assessing recognition in solution is under<br />
investigation and will be published in due course.<br />
spectrometer operating at 400 MHz for 1 H-NMR and 100 MHz<br />
for 13 C-NMR in DMSO-d 6 using a 5 mm probe. <strong>The</strong> chemical<br />
shifts (δ) are reported in parts per million and were referenced to<br />
the residual solvent peak. <strong>The</strong> following abbreviations are used:<br />
s, singlet; m, multiplet; br, broad signal. Mass spectra were<br />
recorded using HCTultra and ESI-TOF Bruker Daltonics mass<br />
spectrometers and samples were dissolved in DMF, acetonitrile<br />
and water using the positive and negative electrospray ionization<br />
modes. Calibration was carried out using a 0.1 M solution <strong>of</strong><br />
sodium formate in the enhanced quadratic mode prior to each<br />
experimental run. <strong>The</strong> results <strong>of</strong> measurements were processed<br />
using Compass 1.3 data analysis s<strong>of</strong>tware for a Bruker Daltonics<br />
time <strong>of</strong> flight mass spectrometer (micrOTOF). Molecular<br />
modeling calculations were carried out with HyperChem<br />
s<strong>of</strong>tware (Release 8.0) at the MM+ levels in vacuo and no influence<br />
<strong>of</strong> solvents was taken into account in these calculations.<br />
41,42 Circular Dichroism measurements were carried out<br />
using Jasco-J-810 Spectropolarimeter in H 2 O and DMSO.<br />
Preloaded wang resin, N,N-diisopropylethylamine (DIEA),<br />
O-(benzotriazol-1-yl)-hydroxybenzotriazole (HOBt), N,N,N',N'-<br />
tetramethyluranium hexafluorophosphate (HBTU), 9-fluorenylmethoxycarbonyl)-amino<br />
acid derivatives (1-Fmoc), DMF, N-<br />
methylpyrrolidone (NMP), trifluoroacetic acid (TFA), 1,2-ethanedithiol<br />
(EDT) and other chemicals required for peptide<br />
synthesis were bought from Iris Biotech GmbH (Marktredwitz,<br />
Germany). Peptides were synthesized with standard solid phase<br />
peptide synthesis technique and it was carried out on an<br />
automated peptide synthesizer (ABI-433A, Applied Biosystems,<br />
Foster city, USA). Fmoc protected preloaded resin was used to<br />
grow peptide chain on it. In detail, deprotection <strong>of</strong> the N-terminal<br />
<strong>of</strong> resin bound amino acid was performed by 20% piperidine in<br />
NMP and the C-terminal <strong>of</strong> other protected amino acid was<br />
activated using HBTU/ HOBt/DIEA (1:1:2) in DMF for coupling<br />
with the free N-terminal <strong>of</strong> the resin bound amino acid. A<br />
mixture <strong>of</strong> 82.5% TFA, 5% phenol, 5% water, 5% thioanisol and<br />
2.5% EDT was used to separate the peptide from resin as well as<br />
to remove the side chain protecting groups. Subsequently, the<br />
peptide was isolated through precipitation in cold (–20 ᴼC)<br />
diethyl ether and lyophilized by Christ freeze dryers (Martin<br />
Christ Gefriertrocknungsanlagen GmbH, Osterode am Harz,<br />
Germany). Peptides were purified by high performance liquid<br />
chromatography (HPLC) and analyzed by HRMS. Dicarbohydrazides<br />
1-3 were synthesized according to literature. 27,28<br />
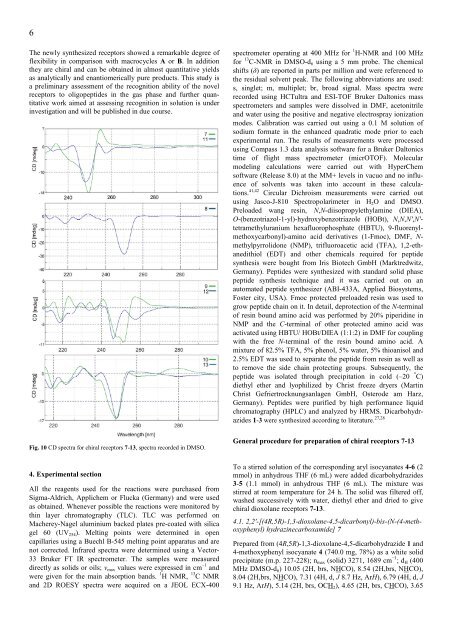
Fig. 10 CD spectra for chiral receptors 7-13, spectra recorded in DMSO.<br />
4. Experimental section<br />
All the reagents used for the reactions were purchased from<br />
Sigma-Aldrich, Applichem or Flucka (Germany) and were used<br />
as obtained. Whenever possible the reactions were monitored by<br />
thin layer chromatography (TLC). TLC was performed on<br />
Macherey-Nagel aluminium backed plates pre-coated with silica<br />
gel 60 (UV 254 ). Melting points were determined in open<br />
capillaries using a Buechl B-545 melting point apparatus and are<br />
not corrected. Infrared spectra were determined using a Vector-<br />
33 Bruker FT IR spectrometer. <strong>The</strong> samples were measured<br />
directly as solids or oils; ν max values were expressed in cm –1 and<br />
were given for the main absorption bands. 1 H NMR, 13 C NMR<br />
and 2D ROESY spectra were acquired on a JEOL ECX-400<br />
General procedure for preparation <strong>of</strong> chiral receptors 7-13<br />
To a stirred solution <strong>of</strong> the corresponding aryl isocyanates 4-6 (2<br />
mmol) in anhydrous THF (6 mL) were added dicarbohydrazides<br />
3-5 (1.1 mmol) in anhydrous THF (6 mL). <strong>The</strong> mixture was<br />
stirred at room temperature for 24 h. <strong>The</strong> solid was filtered <strong>of</strong>f,<br />
washed successively with water, diethyl ether and dried to give<br />
chiral dioxolane receptors 7-13.<br />
4.1. 2,2'-[(4R,5R)-1,3-dioxolane-4,5-dicarbonyl)-bis-(N-(4-methoxyphenyl)<br />
hydrazinecarboxamide] 7<br />
Prepared from (4R,5R)-1,3-dioxolane-4,5-dicarbohydrazide 1 and<br />
4-methoxyphenyl isocyanate 4 (740.0 mg, 78%) as a white solid<br />
precipitate (m.p. 227-228); n max (solid) 3271, 1689 cm −1 ; d H (400<br />
MHz DMSO-d 6 ) 10.05 (2H, brs, NHCO), 8.54 (2H,brs, NHCO),<br />
8.04 (2H,brs, NHCO), 7.31 (4H, d, J 8.7 Hz, ArH), 6.79 (4H, d, J<br />
9.1 Hz, ArH), 5.14 (2H, brs, OCH 2 ), 4.65 (2H, brs, CHCO), 3.65