bbc 2015
BBC2015_booklet
BBC2015_booklet
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
BeNeLux Bioinformatics Conference – Antwerp, December 7-8 <strong>2015</strong><br />
Abstract ID: P<br />
Poster<br />
10th Benelux Bioinformatics Conference <strong>bbc</strong> <strong>2015</strong><br />
P61. THE DETECTION OF PURIFYING SELECTION DURING TUMOUR<br />
EVOLUTION UNVEILS CANCER VULNERABILITIES<br />
Jimmy Van den Eynden 1* & Erik Larsson 1 .<br />
Department of Medical Biochemistry and Cell Biology, Institute of Biomedicine, The Sahlgrenska Academy, University<br />
of Gothenburg, Sweden. * jimmy.van.den.eynden@gu.se<br />
Identification of somatic mutation patterns indicative of positive selection arguably has become the major goal of cancer<br />
genomics. This is motivated by a search for cancer driver genes and pathways that are recurrently activated in tumours<br />
but not normal cells, thus providing possible therapeutic windows. However, cancer cells additionally depend on a large<br />
number of basic cellular processes, and elevated sensitivity to inhibition of certain essential non-driver genes has been<br />
demonstrated in some cases. While such vulnerability genes should in theory be identifiable based on strong purifying<br />
(negative) selection in tumors, these patterns have been elusive and purifying selection remains underexplored in cancer.<br />
We established a new methodology and, using mutational data from 25 TCGA tumor types, we show for the first time<br />
that negative selection in candidate vulnerability genes can be detected.<br />
INTRODUCTION<br />
Recently it was shown that a hemizygous deletion of the<br />
well–known tumour suppressor gene TP53 creates<br />
therapeutic vulnerability in colorectal cancer due to<br />
concomitant loss of the neighbouring gene POLR2A (Liu<br />
et al., <strong>2015</strong>).<br />
As any damaging mutation occurring in the single allele of<br />
a hemizygously deleted essential gene, like POLR2A, is<br />
expected to lead to cell death, we hypothesized that<br />
purifying selection in these genes could be unveiled by<br />
demonstrating a lower number of damaging mutations<br />
then could be expected in the absence of any selection.<br />
Therefore we used the POLR2A case as a proof-ofconcept<br />
to develop a methodology to detect purifying<br />
selection in large genome sequencing datasets.<br />
METHODS<br />
Mutation and copy number data from 25 different cancers<br />
types and 7,871 samples were downloaded from the<br />
TCGA data portal and pooled together in a large pancancer<br />
dataset. Different mutational functional impact<br />
scores were calculated using Annovar. Copy number data<br />
were analyzed using Gistic 2.0 to differentiate POLR2A<br />
copy number neutral from hemizygously deleted samples.<br />
RESULTS & DISCUSSION<br />
POLR2A was found to be hemizygously deleted in 29% of<br />
all samples. As expected, in over 99% this deletion was<br />
part of the TP53 (driving) deletion on chromosome 17.<br />
POLR2A was mutated 228 times in 2.3% of all samples.<br />
While 14 nonsense mutations and small out-of-frame<br />
insertions or deletions occurred in the copy number<br />
neutral group, none of these damaging mutations were<br />
found in the deletion group (p=0.03, fisher test),<br />
suggesting purifying selection against this type of<br />
mutations.<br />
Next to these truncating mutations, also missense<br />
mutations that have a damaging effect on the gene’s<br />
protein function are expected to be selected against.<br />
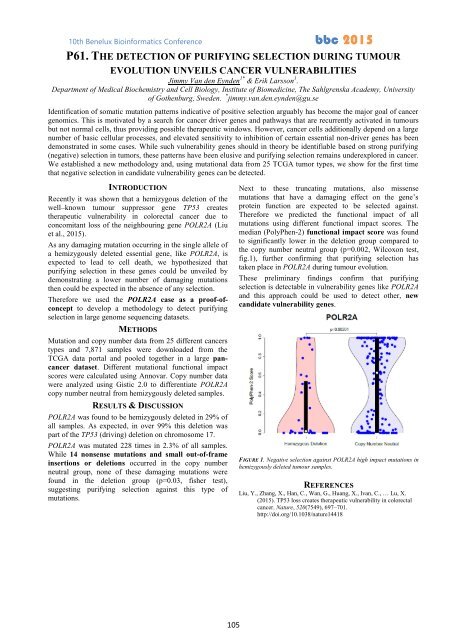
Therefore we predicted the functional impact of all<br />
mutations using different functional impact scores. The<br />
median (PolyPhen-2) functional impact score was found<br />
to significantly lower in the deletion group compared to<br />
the copy number neutral group (p=0.002, Wilcoxon test,<br />
fig.1), further confirming that purifying selection has<br />
taken place in POLR2A during tumour evolution.<br />
These preliminary findings confirm that purifying<br />
selection is detectable in vulnerability genes like POLR2A<br />
and this approach could be used to detect other, new<br />
candidate vulnerability genes.<br />
FIGURE 1. Negative selection against POLR2A high impact mutations in<br />
hemizygously deleted tumour samples.<br />
REFERENCES<br />
Liu, Y., Zhang, X., Han, C., Wan, G., Huang, X., Ivan, C., … Lu, X.<br />
(<strong>2015</strong>). TP53 loss creates therapeutic vulnerability in colorectal<br />
cancer. Nature, 520(7549), 697–701.<br />
http://doi.org/10.1038/nature14418<br />
105