bbc 2015
BBC2015_booklet
BBC2015_booklet
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
BeNeLux Bioinformatics Conference – Antwerp, December 7-8 <strong>2015</strong><br />
Abstract ID: O5<br />
Oral presentation<br />
10th Benelux Bioinformatics Conference <strong>bbc</strong> <strong>2015</strong><br />
O5. INFERRING DEVELOPMENTAL CHRONOLOGIES FROM SINGLE CELL<br />
RNA<br />
Robrecht Cannoodt 1,2,3* , Katleen De Preter 3 & Yvan Saeys 1,2 .<br />
Data Mining and Modelling for Biomedicine group, VIB Inflammation Research Center, Ghent 1 ; Department of<br />
Respiratory Medicine, Ghent University Hospital, Ghent 2 ; Center of Medical Genetics, Ghent University Hospital,<br />
Ghent 3 . * robrecht.cannoodt@ugent.be<br />
With the advent of single cell RNA sequencing, it is now possible to analyse the transcriptomes of hundreds of individual<br />
cells in an unbiased manner. Reconstructing the developmental chronology of differentiating cells is a challenging task,<br />
and doing so in a unsupervised and robust manner is a hitherto untackled problem. We developed a truly unsupervised<br />
developmental chronology inference technique, and evaluated its performance and robustness using multiple datasets.<br />
INTRODUCTION<br />
Early attempts at inferring the chronologies of single cells<br />
are MONOCLE (Trapnell et al., 2014) and NBOR<br />
(Schlitzer et al., <strong>2015</strong>). However, these techniques are not<br />
unsupervised as they require knowledge of the cell type of<br />
each cell prior to analysis, which biases the results to prior<br />
knowledge and possibly obstructs the discovery of novel<br />
subpopulations.<br />
METHODS<br />
Our approach consists of four steps.<br />
In the first step, the feature space (~30000 genes) is<br />
reduced to three dimensions.<br />
Secondly, outliers are detected and removed, using a K-<br />
nearest neighbour approach. After outlier removal, the<br />
original feature space is again reduced to three dimensions.<br />
Next, a nonparametric nonlinear curve is iteratively fitted<br />
to the data.<br />
Finally, each cell is projected onto the curve, thus<br />
resulting in a cell chronology.<br />
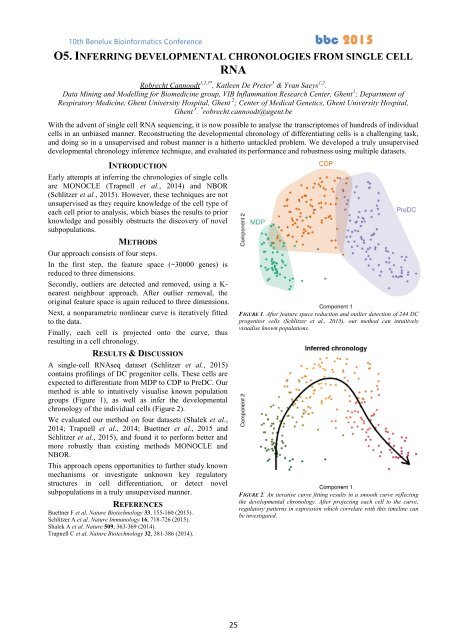
RESULTS & DISCUSSION<br />
A single-cell RNAseq dataset (Schlitzer et al., <strong>2015</strong>)<br />
contains profilings of DC progenitor cells. These cells are<br />
expected to differentiate from MDP to CDP to PreDC. Our<br />
method is able to intuitively visualise known population<br />
groups (Figure 1), as well as infer the developmental<br />
chronology of the individual cells (Figure 2).<br />
We evaluated our method on four datasets (Shalek et al.,<br />
2014; Trapnell et al., 2014; Buettner et al., <strong>2015</strong> and<br />
Schlitzer et al., <strong>2015</strong>), and found it to perform better and<br />
more robustly than existing methods MONOCLE and<br />
NBOR.<br />
This approach opens opportunities to further study known<br />
mechanisms or investigate unknown key regulatory<br />
structures in cell differentiation, or detect novel<br />
subpopulations in a truly unsupervised manner.<br />
REFERENCES<br />
Buettner F et al. Nature Biotechnology 33, 155-160 (<strong>2015</strong>).<br />
Schlitzer A et al. Nature Immunology 16, 718-726 (<strong>2015</strong>).<br />
Shalek A et al. Nature 509, 363-369 (2014).<br />
Trapnell C et al. Nature Biotechnology 32, 381-386 (2014).<br />
FIGURE 1. After feature space reduction and outlier detection of 244 DC<br />
progenitor cells (Schlitzer et al., <strong>2015</strong>), our method can intuitively<br />
visualise known populations.<br />
FIGURE 2. An iterative curve fitting results in a smooth curve reflecting<br />
the developmental chronology. After projecting each cell to the curve,<br />
regulatory patterns in expression which correlate with this timeline can<br />
be investigated.<br />
25