bbc 2015
BBC2015_booklet
BBC2015_booklet
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
BeNeLux Bioinformatics Conference – Antwerp, December 7-8 <strong>2015</strong><br />
Abstract ID: 000 Category: Abstract template<br />
10th Benelux Bioinformatics Conference <strong>bbc</strong> <strong>2015</strong><br />
P27. DETECTING MIXED MYCOBACTERIUM TUBERCULOSIS INFECTION<br />
AND DIFFERENCES IN DRUG SUSCEPTIBILITY WITH WGS DATA<br />
Arlin Keo 1 & Thomas Abeel 1,2,* .<br />
Delft Bioinformatics Lab, Delft University of Technology , Delft, the Netherlands 1 ; Broad Institute of MIT<br />
and Harvard, Cambridge, MA, USA 2 . * t.abeel@ tudelft.nl<br />
Mycobacterium tuberculosis is a bacterial pathogen that causes tuberculosis and infects millions of people. When a<br />
person is infected with more than one distinct strain type of tuberculosis (TB), referred to as mixed infection, diagnosis<br />
and treatment is complicated. Due to difficulty of diagnosis the prevalence of mixed infections among TB patients<br />
remain uncertain. Whole genome sequencing (WGS) yields a great number of single nucleotide polymorphisms (SNPs)<br />
and offers increased resolution to distinguish distinct strains. Here, we present a tool that maps sample reads against 21<br />
bp cluster specific SNP markers to detect putative mixed infections and estimate the frequencies of the present<br />
subpopulations.<br />
INTRODUCTION<br />
Mycobacterium tuberculosis is a clonal, bacterial pathogen<br />
that causes the pulmonary disease tuberculosis (TB), and it<br />
infects and kills millions of people worldwide [1]. The<br />
study of genetic diversity within the M. tuberculosis<br />
complex (MTBC) is complicated by mixed TB infections,<br />
which happens when a person is infected with more than<br />
one distinct strain type of MTBC. This often results in<br />
poor diagnosis and treatment of patients as the bacterial<br />
subpopulation may have undetected differences in drug<br />
susceptibility [2]. A strain typing method should be able to<br />
distinguish closely related strains, to also allow the<br />
detection of a mixed infection at finer resolutions [3]. This<br />
study aims to detect a possible mixed TB infection at<br />
different levels in MTBC and to determine the frequencies<br />
of the present strains based on established tree paths in the<br />
MTBC phylogenetic tree.<br />
METHODS<br />
A global comprehensive dataset of 5992 MTBC strains<br />
was used for analysis, and 226570 SNPs were extracted<br />
from this set to construct a SNP-based phylogenetic tree<br />
with RAxML. In this bifurcating tree, each branch<br />
represents a cluster of strains and splits into two new<br />
monophyletic subclusters of genetically more closely<br />
related strain. These ¨splits¨ were used to define clusters<br />
and subclusters that contain more than 10. Global SNP<br />
association was done for each cluster to get clusterspecific<br />
SNPs, those for which the true positive rate, true<br />
negative rate, positive predictive value, and negative<br />
predictive value were >0.95. Markers were generated from<br />
these SNPs by extending them with 10 bp sequence on<br />
each side based on reference genome H37Rv. Each<br />
hierarchical cluster now has a set of specific SNP markers.<br />
By mapping sample reads against these 21 bp clusterspecific<br />
SNP markers the tool determines the presence of<br />
paths in the phylogenetic tree that start at the MTBC root<br />
node. Paths that split indicate the presence of multiple<br />
strains and thus a mixed infection.<br />
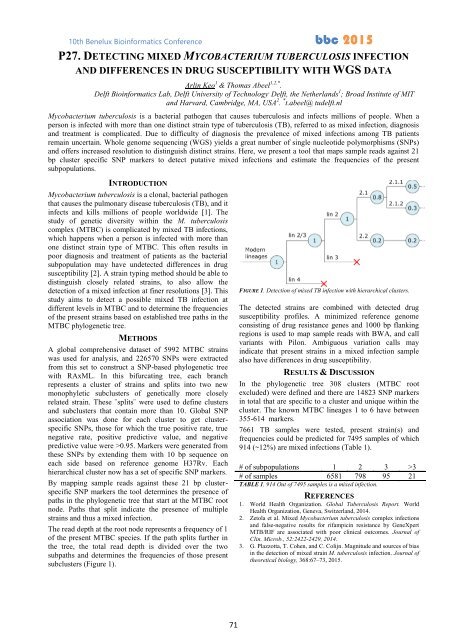
The read depth at the root node represents a frequency of 1<br />
of the present MTBC species. If the path splits further in<br />
the tree, the total read depth is divided over the two<br />
subpaths and determines the frequencies of those present<br />
subclusters (Figure 1).<br />
FIGURE 1. Detection of mixed TB infection with hierarchical clusters.<br />
The detected strains are combined with detected drug<br />
susceptibility profiles. A minimized reference genome<br />
consisting of drug resistance genes and 1000 bp flanking<br />
regions is used to map sample reads with BWA, and call<br />
variants with Pilon. Ambiguous variation calls may<br />
indicate that present strains in a mixed infection sample<br />
also have differences in drug susceptibility.<br />
RESULTS & DISCUSSION<br />
In the phylogenetic tree 308 clusters (MTBC root<br />
excluded) were defined and there are 14823 SNP markers<br />
in total that are specific to a cluster and unique within the<br />
cluster. The known MTBC lineages 1 to 6 have between<br />
355-614 markers.<br />
7661 TB samples were tested, present strain(s) and<br />
frequencies could be predicted for 7495 samples of which<br />
914 (~12%) are mixed infections (Table 1).<br />
# of subpopulations 1 2 3 >3<br />
# of samples 6581 798 95 21<br />
TABLE 1. 914 Out of 7495 samples is a mixed infection.<br />
REFERENCES<br />
1. World Health Organization. Global Tuberculosis Report. World<br />
Health Organization, Geneva, Switzerland, 2014.<br />
2. Zetola et al. Mixed Mycobacterium tuberculosis complex infections<br />
and false-negative results for rifampicin resistance by GeneXpert<br />
MTB/RIF are associated with poor clinical outcomes. Journal of<br />
Clin. Microb., 52:2422-2429, 2014.<br />
3. G. Plazzotta, T. Cohen, and C. Colijn. Magnitude and sources of bias<br />
in the detection of mixed strain M. tuberculosis infection. Journal of<br />
theoretical biology, 368:67–73, <strong>2015</strong>.<br />
71