bbc 2015
BBC2015_booklet
BBC2015_booklet
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
BeNeLux Bioinformatics Conference – Antwerp, December 7-8 <strong>2015</strong><br />
Abstract ID: P<br />
Poster<br />
10th Benelux Bioinformatics Conference <strong>bbc</strong> <strong>2015</strong><br />
P1. KNN-MDR APPROACH FOR DETECTING GENE-GENE<br />
INTERACTIONS<br />
Sinan Abo alchamlat 1 & Frédéric Farnir 1,* .<br />
Fundamental and Applied Research for Animals & Health (FARAH), Sustainable Animal Production, University of<br />
Liège 1 . * f.farnir@ulg.ac.be<br />
These last years have seen the emergence of a wealth of biological information. Facilitated access to the genome<br />
sequence, along with massive data on genes expression and on proteins have revolutionized the research in many fields<br />
of biology. For example, the identification of up to several millions SNPs in many species and the development of chips<br />
allowing for an effective genotyping of these SNPs in large cohorts have triggered the need for statistical models able to<br />
identify the effects of individual and of interacting SNPs on phenotypic traits in this new high-dimensional landscape.<br />
Our work is a contribution to this field...............................................................................................................<br />
INTRODUCTION<br />
GWAS has allowed the identification of hundreds of<br />
genetic variants associated to complex diseases and traits,<br />
and provided valuable information into their genetic<br />
architecture (Wu M et al., 2010). Nevertheless, most<br />
variants identified so far have been found to confer<br />
relatively small information about the relationship<br />
between changes at the genomic level and phenotypes<br />
because of the lack of reproducibility of the findings, or<br />
because these variants most of the time explain only a<br />
small proportion of the underlying genetic variation (Fang<br />
G et al., 2012). This observation, quoted as the ‘missing<br />
heritability’ problem (Manolio T et al., 2009) of course<br />
raises the question: where does the unexplained genetic<br />
variation come from? A tentative explanation is that genes<br />
do not work in isolation, leading to the idea that sets of<br />
genes (or genes networks) could have a major effect on the<br />
tested traits while almost no marginal – i.e. individual<br />
gene – effect is detectable. Consequently, an important<br />
question concerns the exact relationship between the<br />
genomic configuration, including the interactions between<br />
the involved genes, and the phenotypic expression.<br />
METHODS<br />
To tackle this subject, different statistical methods such as<br />
MDR (Multi Dimensional Reduction) have been proposed<br />
for detecting gene-gene interaction (Ritchie, D., et al.,<br />
2001); their relative performances remain largely unclear,<br />
and their extension to situations combining many variants<br />
turns out to be challenging. So we propose a novel MDR<br />
approach using K-Nearest Neighbors (KNN) methodology<br />
(KNN-MDR) for detecting gene-gene interaction as a<br />
possible alternative, especially when the number of<br />
involved determinants is potentially high. The idea behind<br />
our method is to replace the status allocation used in<br />
classical MDR methods by a KNN approach: the majority<br />
vote occurs in the k (a parameter that must be tuned and<br />
depends on the various possible scenarios) nearest<br />
neighbors instead of within the (potentially empty) cell<br />
determined by the tested attributes of the individual to be<br />
classified. The steps other than classification are identical<br />
in both methods (i.e. cross-validation, attributes selection,<br />
training and tests balanced accuracy computations, best<br />
model selection procedure).<br />
RESULTS & DISCUSSION<br />
Experimental results on both simulated data and real<br />
genome-wide data from Wellcome Trust Case Control<br />
Consortium (WTCCC) (Wellcome Trust Case Control C.,<br />
2007) show that KNN-MDR has interesting properties in<br />
terms of accuracy and power, and that, in many cases, it<br />
significantly outperforms its recent competitors.<br />
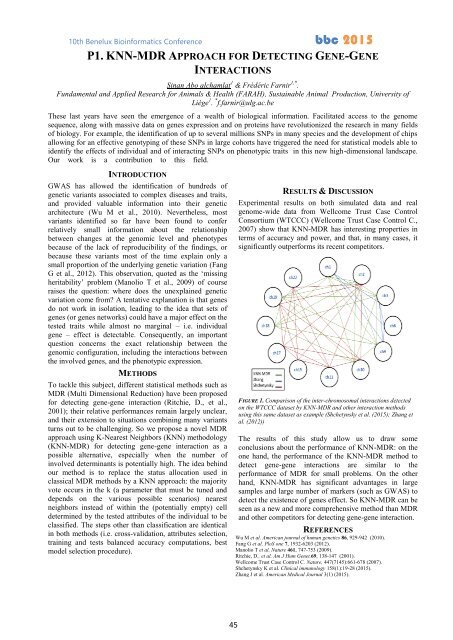
FIGURE 1. Comparison of the inter-chromosomal interactions detected<br />
on the WTCCC dataset by KNN-MDR and other interaction methods<br />
using this same dataset as example (Shchetynsky et al. (<strong>2015</strong>); Zhang et<br />
al. (2012))<br />
The results of this study allow us to draw some<br />
conclusions about the performance of KNN-MDR: on the<br />
one hand, the performance of the KNN-MDR method to<br />
detect gene-gene interactions are similar to the<br />
performance of MDR for small problems. On the other<br />
hand, KNN-MDR has significant advantages in large<br />
samples and large number of markers (such as GWAS) to<br />
detect the existence of genes effect. So KNN-MDR can be<br />
seen as a new and more comprehensive method than MDR<br />
and other competitors for detecting gene-gene interaction.<br />
REFERENCES<br />
Wu M et al. American journal of human genetics 86, 929-942 (2010).<br />
Fang G et al. PloS one 7, 1932-6203 (2012).<br />
Manolio T et al. Nature 461, 747-753 (2009).<br />
Ritchie, D., et al. Am J Hum Genet,69, 138-147 (2001).<br />
Wellcome Trust Case Control C. Nature, 447(7145):661-678 (2007).<br />
Shchetynsky K et al. Clinical immunology 158(1):19-28 (<strong>2015</strong>).<br />
Zhang J et al. American Medical Journal 3(1) (<strong>2015</strong>).<br />
45