bbc 2015
BBC2015_booklet
BBC2015_booklet
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
BeNeLux Bioinformatics Conference – Antwerp, December 7-8 <strong>2015</strong><br />
Abstract ID: P<br />
Poster<br />
10th Benelux Bioinformatics Conference <strong>bbc</strong> <strong>2015</strong><br />
P4. DISEASE-SPECIFIC NETWORK CONSTRUCTION BY SEED-AND-EXTEND<br />
Ganna Androsova 1* , Reinhard Schneider 1 & Roland Krause 1 .<br />
Luxembourg Centre for Systems Biomedicine, University of Luxembourg, Belval, Luxembourg 1 .<br />
* ganna.androsova@uni.lu<br />
INTRODUCTION<br />
Molecular interaction networks are dense structures of<br />
protein interactions, from which we would like to extract<br />
relevant sub-networks specific to the disease of interest.<br />
Such a disease-specific network is often constructed by the<br />
seed-and-extend algorithm, which extracts the relevant<br />
genes from an organism-wide, weighted interaction<br />
network, typically as its first-neighbourhood. Seed-andextend<br />
is suitable when disease biomarkers are poorly<br />
investigated and the knowledge about biomarker<br />
interaction partners is missing or when the interacting<br />
partners are established but the connections are missing<br />
between them.<br />
Our syndrome of interest is the postoperative cognitive<br />
impairment frequently experienced by elderly patients,<br />
characterized by progressive cognitive and sensory decline.<br />
The acute phase of cognitive impairment is postoperative<br />
delirium (POD). The underlying pathophysiological<br />
mechanisms have not been studied in depth due to<br />
mulitifactorial pathogenesis of this postoperative cognitive<br />
impairment. The known POD-related genes can be<br />
integrated into the draft network for exploration on a<br />
systems level.<br />
Here, we investigate how stable the results of such<br />
analysis are when the input set of seed genes is varied, and<br />
what is the role of stringency in the initial selection of the<br />
networks. Ideally, we would like to find the “sweet spot”<br />
that provides a biologically meaningful trade-off between<br />
false-positives and -negatives to be used for such analyses.<br />
METHODS<br />
The list of disease-related genes/proteins was retrieved<br />
from literature studies in the PubMed database.<br />
We extended the seed list with directly linked interactors<br />
by seed-and-extend from protein-protein interaction<br />
network databases. We extracted all interactions between<br />
seeds and connected neighbours, which resulted in the<br />
first-degree network.<br />
Next, we evaluated a biological enrichment of the<br />
extracted network, its topological parameters, overlap with<br />
other diseases and clustered the network into the smaller<br />
sub-networks.<br />
RESULTS & DISCUSSION<br />
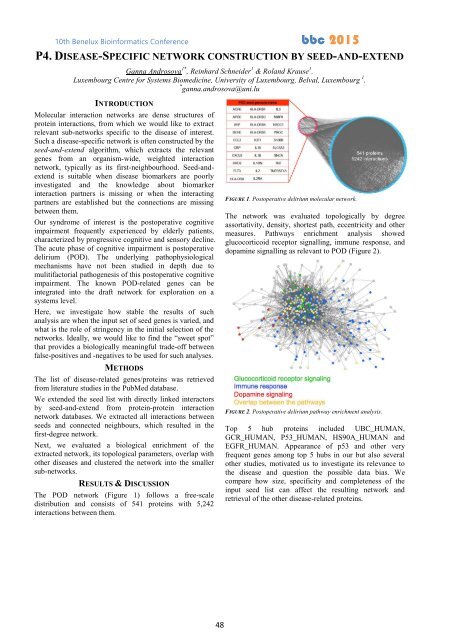
The POD network (Figure 1) follows a free-scale<br />
distribution and consists of 541 proteins with 5,242<br />
interactions between them.<br />
FIGURE 1. Postoperative delirium molecular network.<br />
The network was evaluated topologically by degree<br />
assortativity, density, shortest path, eccentricity and other<br />
measures. Pathways enrichment analysis showed<br />
glucocorticoid receptor signalling, immune response, and<br />
dopamine signalling as relevant to POD (Figure 2).<br />
FIGURE 2. Postoperative delirium pathway enrichment analysis.<br />
Top 5 hub proteins included UBC_HUMAN,<br />
GCR_HUMAN, P53_HUMAN, HS90A_HUMAN and<br />
EGFR_HUMAN. Appearance of p53 and other very<br />
frequent genes among top 5 hubs in our but also several<br />
other studies, motivated us to investigate its relevance to<br />
the disease and question the possible data bias. We<br />
compare how size, specificity and completeness of the<br />
input seed list can affect the resulting network and<br />
retrieval of the other disease-related proteins.<br />
48