bbc 2015
BBC2015_booklet
BBC2015_booklet
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
BeNeLux Bioinformatics Conference – Antwerp, December 7-8 <strong>2015</strong><br />
Abstract ID: P<br />
Poster<br />
10th Benelux Bioinformatics Conference <strong>bbc</strong> <strong>2015</strong><br />
P34. FUNCTIONAL SUBGRAPH ENRICHMENTS<br />
FOR NODE SETS IN REGULATORY NETWORKS<br />
Pieter Meysman 1,2* , Yvan Saeys 3,4 , Ehsan Sabaghian 5,6 , Wout Bittremieux 1,2 ,<br />
Yves van de Peer 5,6 , Bart Goethals 1 & Kris Laukens 1,2 .<br />
Advanced Database Research and Modeling (ADReM), University of Antwerp 1 ; Biomedical informatics research center<br />
Antwerpen (biomina) 2 ; VIB Inflammation Research Center 3 ; Department of Respiratory Medicine, Ghent University 4 ;<br />
Department of Plant Biotechnology and Bioinformatics, Ghent University 5 ; Department of Plant Systems Biology,<br />
VIB/Ghent University 6 . * pieter.meysman@uantwerpen.be<br />
We have developed a subgroup discovery algorithm to find subgraphs in a single graph that are associated with a given<br />
set of nodes. The association between a subgraph pattern and a set of vertices is defined by its significant enrichment<br />
based on a Bonferroni-corrected hypergeometric probability value, and can therefore be considered as a network-focused<br />
extension of traditional gene ontology enrichment analysis. We demonstrate the operation of this algorithm by applying it<br />
on two transcriptional regulatory networks and show that we can find relevant functional subgraphs enriched for the<br />
selected nodes.<br />
INTRODUCTION<br />
Frequent subgraph mining (FSM) is a common but<br />
complex problem within the data mining field that has<br />
gained in importance as more graph data has become<br />
available. However traditional FSM finds all frequent<br />
subgraphs within the graph dataset, while often a more<br />
interesting query is to find the subgraphs that are most<br />
associated with a specific set of nodes. Nodes of interest<br />
might be those that are associated with a specific disease,<br />
or those that are differentially expressed in an omics<br />
experiment.<br />
METHODS<br />
To address this issue, we developed a novel subgraph<br />
mining algorithm that can efficiently construct, match and<br />
test candidate subgraphs against the given graph for<br />
enrichment within a specific set of nodes (Meysman et al.<br />
<strong>2015</strong>). To allow the enrichment testing, each candidate<br />
subgraph is built around a ‘source’ node. A subgraph<br />
match where the source node corresponds to a node of<br />
interest is counted as a ‘hit’. If the source node is not a<br />
node of interest, it is counted as a background hit. In this<br />
manner the problem of enrichment can be easily tested<br />
using a hypergeometric test. Furthermore, we show that<br />
this definition of enrichment allows us to drastically prune<br />
the search space that the algorithm must traverse to find all<br />
enriched subgraphs.<br />
An implementation of the algorithm is available at<br />
http://adrem.ua.ac.be/sigsubgraph.<br />
RESULTS & DISCUSSION<br />
The first data set concerned the yeast genes that have<br />
remained in duplicate following the most recent whole<br />
genome duplication. Within the yeast transcriptional<br />
network, we found that these duplicate genes were<br />
enriched for self-regulating motifs (e.g. feedback loops,<br />
self edges, etc.), which matches the duplicated nature of<br />
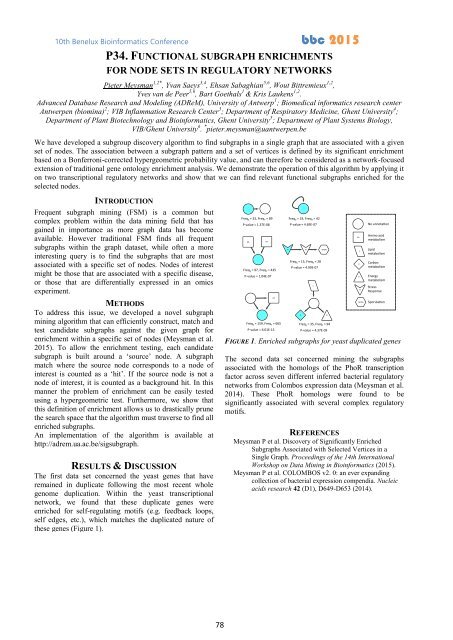
these genes (Figure 1).<br />
FIGURE 1. Enriched subgraphs for yeast duplicated genes<br />
The second data set concerned mining the subgraphs<br />
associated with the homologs of the PhoR transcription<br />
factor across seven different inferred bacterial regulatory<br />
networks from Colombos expression data (Meysman et al.<br />
2014). These PhoR homologs were found to be<br />
significantly associated with several complex regulatory<br />
motifs.<br />
REFERENCES<br />
Meysman P et al. Discovery of Significantly Enriched<br />
Subgraphs Associated with Selected Vertices in a<br />
Single Graph. Proceedings of the 14th International<br />
Workshop on Data Mining in Bioinformatics (<strong>2015</strong>).<br />
Meysman P et al. COLOMBOS v2. 0: an ever expanding<br />
collection of bacterial expression compendia. Nucleic<br />
acids research 42 (D1), D649-D653 (2014).<br />
78