bbc 2015
BBC2015_booklet
BBC2015_booklet
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
BeNeLux Bioinformatics Conference – Antwerp, December 7-8 <strong>2015</strong><br />
Abstract ID: O17<br />
Oral presentation<br />
10th Benelux Bioinformatics Conference <strong>bbc</strong> <strong>2015</strong><br />
O17. GENE CO-EXPRESSION ANALYSIS IDENTIFIES BRAIN REGIONS AND<br />
CELL TYPES INVOLVED IN MIGRAINE PATHOPHYSIOLOGY: A GWAS-<br />
BASED STUDY USING THE ALLEN HUMAN BRAIN ATLAS<br />
Sjoerd M.H. Huisman 1,2* , Else Eising 3 , Ahmed Mahfouz 1,2 , Lisanne Vijfhuizen 3 , International Headache Genetics<br />
Consortium, Boudewijn P.F. Lelieveldt 2 , Arn M.J.M. van den Maagdenberg 3,4 & Marcel J.T. Reinders 1 .<br />
DBL, Dept. of Intelligent Systems, Delft University of Technology, The Netherlands 1 ; LKEB, Dept. of Radiology, Leiden<br />
University Medical Center, The Netherlands 2 ; Dept. of Human Genetics, Leiden University Medical Center, The<br />
Netherlands 3 ; Dept. of Neurology, Leiden University Medical Center, The Netherlands 4 . * s.m.h.huisman@tudelft.nl<br />
Migraine is a common brain disorder, with a heritability of around 50%. To understand the genetic component of this<br />
disease, a large genome wide association study has been carried out. Several loci were identified, but their interpretation<br />
remained challenging. We integrated the GWAS results with gene expression data, from healthy human brains, to<br />
identify anatomical regions and biological pathways implicated in migraine pathophysiology.<br />
INTRODUCTION<br />
Genome Wide Association Studies (GWAS) are<br />
frequently used to find common variants with small effect<br />
sizes. However, they often provide researchers with short<br />
lists of single nucleotide polymorphisms (SNPs) with<br />
uncertain connections to biological functions.<br />
We present an analysis of GWAS data for migraine, where<br />
the full list of SNP statistics is used to find groups of<br />
functionally related migraine-associated genes. For this<br />
end we make use of gene co-expression in the healthy<br />
human brain.<br />
We performed genome wide clustering of genes, followed<br />
by enrichment analysis for migraine candidate genes. In<br />
addition, we constructed local co-expression networks<br />
around high-confidence genes. Both approaches converge<br />
on distinct biological functions and brain regions of<br />
interest.<br />
METHODS<br />
Migraine GWAS data was obtained from the International<br />
Headache Genetics Consortium, with 23,285 cases and<br />
95,425 controls (Anttila et al., 2013). Genes were scored<br />
by SNP load and divided into high-confidence genes,<br />
migraine candidate genes, and non-migraine genes.<br />
Spatial gene expression data in the healthy adult human<br />
brain was obtained from the Allen Brain Institute<br />
(Hawrylycz et al., 2012). It contains microarray<br />
expression values of 3702 samples from 6 donors. Robust<br />
gene co-expressions were used to cluster genes into 18<br />
modules, which were then tested for enrichment of<br />
migraine candidate genes, and functionally characterized.<br />
In a second approach, local co-expression networks were<br />
built around the high-confidence migraine genes. These<br />
local networks were then compared to the modules of the<br />
first approach.<br />
RESULTS & DISCUSSION<br />
The genome wide analysis revealed several modules of<br />
genes enriched in migraine candidates. Two modules have<br />
preferential expression in the cerebral cortex and are<br />
enriched in synapse related annotations and neuron<br />
specific genes. A third module contains oligodendrocytes<br />
and genes preferentially expressed in subcortical regions.<br />
The local co-expression networks, of the second approach,<br />
converge on the same pathways and expression patterns,<br />
even though the high confidence genes lie mostly outside<br />
of the modules of interest. This provides a control to the<br />
results of the first approach.<br />
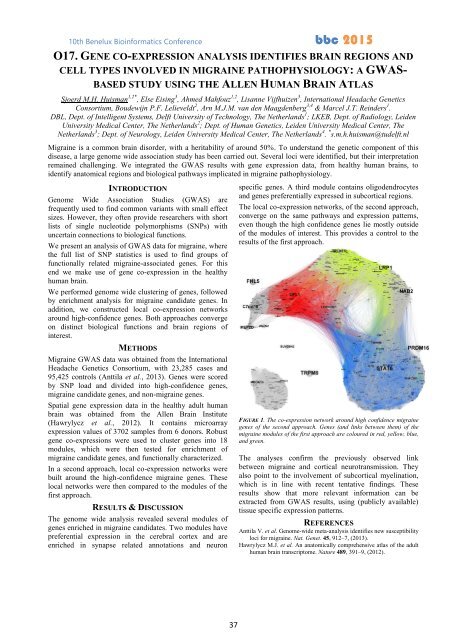
FIGURE 1. The co-expression network around high confidence migraine<br />
genes of the second approach. Genes (and links between them) of the<br />
migraine modules of the first approach are coloured in red, yellow, blue,<br />
and green.<br />
The analyses confirm the previously observed link<br />
between migraine and cortical neurotransmission. They<br />
also point to the involvement of subcortical myelination,<br />
which is in line with recent tentative findings. These<br />
results show that more relevant information can be<br />
extracted from GWAS results, using (publicly available)<br />
tissue specific expression patterns.<br />
REFERENCES<br />
Anttila V. et al. Genome-wide meta-analysis identifies new susceptibility<br />
loci for migraine. Nat. Genet. 45, 912–7, (2013).<br />
Hawrylycz M.J. et al. An anatomically comprehensive atlas of the adult<br />
human brain transcriptome. Nature 489, 391–9, (2012).<br />
37