bbc 2015
BBC2015_booklet
BBC2015_booklet
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
BeNeLux Bioinformatics Conference – Antwerp, December 7-8 <strong>2015</strong><br />
Abstract ID: P<br />
Poster<br />
10th Benelux Bioinformatics Conference <strong>bbc</strong> <strong>2015</strong><br />
P67. IDENTIFICATION OF ANTIBIOTIC RESISTANCE MECHANISMS USING<br />
A NETWORK-BASED APPROACH<br />
Bram Weytjens 1,2,3,4 , Dries De Maeyer 1,2,,3,4 & Kathleen Marchal 1,2,4 *.<br />
Dept. of Information Technology (INTEC, iMINDS), UGent, Ghent, 9052, Belgium 1 ; Dept. of Plant Biotechnology and<br />
Bioinformatics, Ghent University, Technologiepark 927, 9052 Gent, Belgium 2 ; Dept. of Microbial and Molecular<br />
Systems, KU Leuven, Kasteelpark Arenberg 20, B-3001 Leuven, Belgium 3 , Bioinformatics Institute Ghent, Ghent<br />
University, Ghent B-9000, Belgium 4 . * kathleen.marchal@intec.ugent.be<br />
Antibiotic resistance is a growing public health concern as the effectiveness of multiple types of antibiotics is decreasing.<br />
To prevent and combat the further spread of antibiotic resistance in bacteria there is the need to better understand the<br />
relationship between genetic alterations and the (molecular) phenotype of antibiotic resistant strains. As several (-omics)<br />
experiments regarding the attainment of antibiotic resistance by bacteria have already been performed and are publicly<br />
available, we re-analysed a laboratory evolution experiment by Suzuki et al. (Suzuki, 2014) in order to demonstrate the<br />
power of a network-based approach in identifying mutations and molecular pathways driving the resistance phenotype.<br />
INTRODUCTION<br />
While network-based approaches are no longer new in<br />
high-throughput (-omics) analysis, they are not yet widely<br />
used in standard analysis pipelines. We analysed a dataset<br />
consisting of multiple E. coli MDS42 strains, each<br />
independently evolved in the presence of a specific<br />
antibiotic (10 in total). By adapting PheNetic (De Maeyer.<br />
2013), an algorithm which connects genetic alterations to<br />
their differentially expressed genes over a genome-wide<br />
interaction network, we were able to automatically<br />
identify mutations in genes which are known to induce<br />
antibiotic resistance.<br />
METHODS<br />
For every strain whole-genome sequencing data and<br />
microarray data (eQTL data) was available. By finding the<br />
most probable connections between the mutations of every<br />
strain and the strain’s respective expression data over a<br />
biological network, PheNetic was able to not only uncover<br />
potential driver genes and molecular pathways for the<br />
resistance phenotype but also to prioritize the identified<br />
mutations based on the likelihood that they are truly<br />
driving the resistance phenotype. Such network-based<br />
approach has following advantages:<br />
<br />
<br />
Integration of interactomics (network), genomics<br />
and interactomics data<br />
Multiple related datasets can be analyzed together<br />
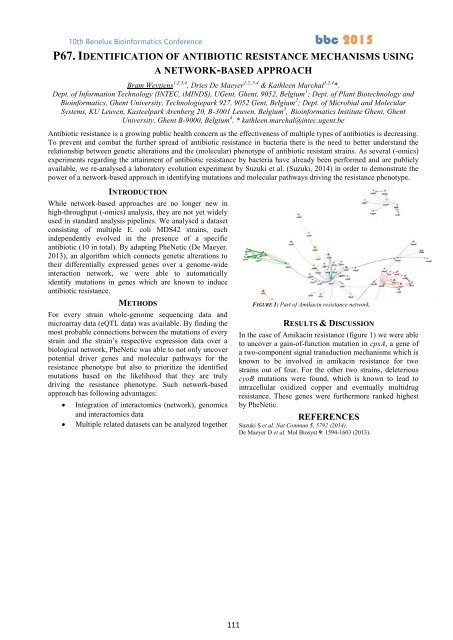
FIGURE 1: Part of Amikacin resistance network.<br />
RESULTS & DISCUSSION<br />
In the case of Amikacin resistance (figure 1) we were able<br />
to uncover a gain-of-function mutation in cpxA, a gene of<br />
a two-component signal transduction mechanisms which is<br />
known to be involved in amikacin resistance for two<br />
strains out of four. For the other two strains, deleterious<br />
cyoB mutations were found, which is known to lead to<br />
intracellular oxidized copper and eventually multidrug<br />
resistance. These genes were furthermore ranked highest<br />
by PheNetic.<br />
REFERENCES<br />
Suzuki S et al. Nat Commun 5, 5792 (2014).<br />
De Maeyer D et al. Mol Biosyst 9: 1594-1603 (2013).<br />
111