- Page 1 and 2:
IMPACT OF HOST PLANT XYLEM FLUID ON
- Page 3 and 4:
0.18 600 Optical density 0.16 0.14
- Page 5 and 6:
OPTIMIZING MARKER-ASSISTED SELECTIO
- Page 7 and 8:
esistant and susceptible genotypes
- Page 9 and 10:
MAP BASED IDENTIFICATION AND POSITI
- Page 11 and 12:
esistant genotypes are in process a
- Page 13 and 14:
BREEDING PIERCE’S DISEASE RESISTA
- Page 15 and 16:
Progeny from crosses of field resis
- Page 17 and 18:
a = (1=low, 4= high); b = (1=green,
- Page 19 and 20:
- 78 -
- Page 21 and 22:
- 80 -
- Page 23 and 24:
v.8) for differences due to the pla
- Page 25 and 26:
SHARPSHOOTER FEEDING BEHAVIOR IN RE
- Page 27 and 28:
correlated with all other findings
- Page 29 and 30:
EFFECTS OF FEEDING SUBSTRATE ON RET
- Page 31 and 32:
REFERENCES Bextine, B. and T. Mille
- Page 33 and 34:
will also provide insights into the
- Page 35 and 36:
OBJECTIVES 1. Determine glassy-wing
- Page 37 and 38:
GWSS per 30 s sweep (seasonal avera
- Page 39 and 40:
ELISA for the presence of GWSS egg
- Page 41 and 42:
Project Leader: Thomas P. Freeman E
- Page 43 and 44:
Freeman, T. P., R. A. Leopold, D. R
- Page 45 and 46:
pathogen, when they move into viney
- Page 47 and 48:
A NOVEL IMMUNOLOGICAL APPROACH FOR
- Page 49 and 50:
1.6 GWSS Adults That Fed on Protein
- Page 51 and 52:
Hagler, J.R. & S.E. Naranjo. 2004.
- Page 53 and 54:
RESULTS: Oviposition Survey Wild gr
- Page 55 and 56:
Host specificity testing: No-choice
- Page 57 and 58:
currently employed morphological ch
- Page 59 and 60:
increase our present understanding
- Page 61 and 62:
BIOLOGY AND MORPHOMETRIC ANALYSIS O
- Page 63 and 64:
Table 1. Mean a developmental durat
- Page 65 and 66:
EFFECTS OF USING CONSTANT AND CYCLI
- Page 67 and 68:
REFERENCES Gautam, R. D. 1986. Effe
- Page 69 and 70:
PARASITISM OF THE GLASSY-WINGED SHA
- Page 71 and 72:
Table 1. Parasitism by G.ashmeadi o
- Page 73 and 74:
Project Leader: Robert F. Luck Dept
- Page 75 and 76:
A more interesting analysis using t
- Page 77 and 78:
MYCOPATHOGENS AND THEIR EXOTOXINS I
- Page 79 and 80:
POPULATION DYNAMICS AND INTERACTION
- Page 81 and 82:
No egg masses were recorded on olea
- Page 83 and 84:
EXPLORATION FOR FACULTATIVE ENDOSYM
- Page 85 and 86:
Although Baumannia and Wolbachia we
- Page 87 and 88:
EFFECTS OF SUBLETHAL DOSES OF IMIDA
- Page 89 and 90:
Table 2. Mortality and flight perfo
- Page 91 and 92:
A NOVEL METHOD TO INDUCE OVIPOSITIO
- Page 93 and 94:
REFERENCES Brodbeck, B. V., P. C. A
- Page 95 and 96:
Cohorts H. coagulata neonates were
- Page 97 and 98:
REFERENCES Blua, M. J. and D. J. W.
- Page 99 and 100:
Figure 2 shows the average numbers
- Page 101 and 102:
REPRODUCTIVE BIOLOGY AND PHYSIOLOGY
- Page 103 and 104:
REFERENCES Blua, M. J., Phillips, P
- Page 105 and 106:
- 164 -
- Page 107 and 108:
- 166 -
- Page 109 and 110:
RESULTS Some plant families had no
- Page 111 and 112: Table 5. Winter, spring and summer
- Page 113 and 114: 16S rDNA is by far the most sequenc
- Page 115 and 116: DNA MICROARRAY AND MUTATIONAL ANALY
- Page 117 and 118: Inhibition of biofilm is BSA concen
- Page 119 and 120: CULTURE-INDEPENDENT ANALYSIS OF END
- Page 121 and 122: Borneman, J., M. Chrobak, G. Della
- Page 123 and 124: P (CFU P OBJECTIVE 1. Determine the
- Page 125 and 126: Purcell, AH and SR Saunders. 1999a.
- Page 127 and 128: Figure 1: Southern blot of tolC::ap
- Page 129 and 130: Project Leader: David Gilchrist Dep
- Page 131 and 132: III. Production of transgenic plant
- Page 133 and 134: RESULTS Construction of cDNA Librar
- Page 135 and 136: CONCLUSIONS Genetic resistance and
- Page 137 and 138: Mutagenesis of Xylella The EZ::TN T
- Page 139 and 140: ISOLATION OF BACTERIOPHAGES SPECIFI
- Page 141 and 142: Project Leader: Michele M. Igo Sect
- Page 143 and 144: outer membrane fractions using two-
- Page 145 and 146: 1995; Purcell and Saunders, 1999).
- Page 147 and 148: DEVELOPMENT OF SSR MARKERS FOR GENO
- Page 149 and 150: Strain Name Host of Origin County o
- Page 151 and 152: ROLE OF ATTACHMENT OF XYLELLA FASTI
- Page 153 and 154: wild-type cells was not conclusive
- Page 155 and 156: DETERMINATION OF GENES CONFERRING H
- Page 157 and 158: S1 S2 S3 S4 Nb123456789bb123456789b
- Page 159 and 160: MULTILOCUS SEQUENCE TYPING TO IDENT
- Page 161: GENOME-WIDE IDENTIFICATION OF RAPID
- Page 165 and 166: EFFECTS OF CHEMICAL MILIEU ON ATTAC
- Page 167 and 168: CONCLUSIONS Our overall objective i
- Page 169 and 170: RESULTS Objective 1. We conducted t
- Page 171 and 172: Redak, R. A., Purcell, A. H., Lopes
- Page 173 and 174: In parallel, an “activating trans
- Page 175 and 176: PATTERNS OF XYLELLA FASTIDIOSA INFE
- Page 177 and 178: % of vessels 100 80 60 40 20 Mugwor
- Page 179 and 180: DOCUMENTATION AND CHARACTERIZATION
- Page 181 and 182: Magnolia002 showed more identity (9
- Page 183 and 184: PLASMID ADDICTION AS A NOVEL APPROA
- Page 185 and 186: GENETIC VARIABILITY OF XYLELLA FAST
- Page 187 and 188: - 246 -
- Page 189 and 190: probing activities). In preliminary
- Page 191 and 192: the slow release valve was opened a
- Page 193 and 194: 12. Hopkins DL (1977) Diseases caus
- Page 195 and 196: The almost complete absence of info
- Page 197 and 198: QUANTIFYING LANDSCAPE-SCALE MOVEMEN
- Page 199 and 200: Table 1. The mean (±SD) ELISA read
- Page 201 and 202: EPIDEMIOLOGICAL ASSESSMENTS OF PIER
- Page 203 and 204: multiply to relatively high (easily
- Page 205 and 206: SPATIAL DATABASE CREATION AND MAINT
- Page 207 and 208: Project Leader: Thomas M. Perring D
- Page 209 and 210: Figure 2. Individual leaves from a
- Page 211 and 212: The state variables, process functi
- Page 213 and 214:
DEVELOPMENT OF A FIELD SAMPLING PLA
- Page 215 and 216:
Figure 1. Three main dispersion pat
- Page 217 and 218:
- 276 -
- Page 219 and 220:
- 278 -
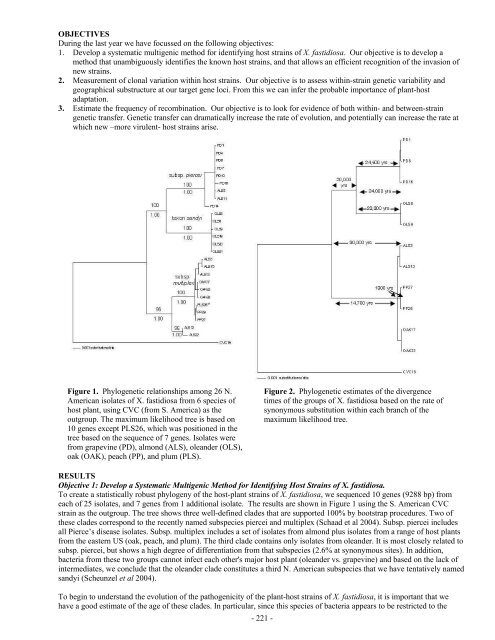
- Page 221 and 222:
OBJECTIVES 1. Track the movement of
- Page 223 and 224:
REFERENCES 1. Beard, C.B., Cordon-R
- Page 225 and 226:
cases, positive phloem samples were
- Page 227 and 228:
EXPLOITING XYLELLA FASTIDIOSA PROTE
- Page 229 and 230:
Figure 2. Polymer disk accumulation
- Page 231 and 232:
CHARACTERIZATION OF NEONICOTINOIDS
- Page 233 and 234:
EVALUATION OF RESISTANCE POTENTIAL
- Page 235 and 236:
Project Leader: Doug Cook Dept. of
- Page 237 and 238:
Based on the in silico analysis, de
- Page 239 and 240:
CONTROL OF PIERCE’S DISEASE THROU
- Page 241 and 242:
Figure 1. Oleander ‘White’ afte
- Page 243 and 244:
RESULTS During the reporting period
- Page 245 and 246:
DEVELOPMENT OF AN ARTIFICIAL DIET A
- Page 247 and 248:
DESIGN OF CHIMERIC ANTI-MICROBIAL P
- Page 249 and 250:
Neutrophil elastase Cecropin B Chim
- Page 251 and 252:
and WTXb is very low (0.00-0.40%) a
- Page 253 and 254:
100 100 0.056 North America 100 0.0
- Page 255 and 256:
GENETIC DIFFERENTIATION AMONG GEOGR
- Page 257 and 258:
Table 1. Single-populations descrip
- Page 259 and 260:
MOLECULAR DISTINCTION BETWEEN POPUL
- Page 261 and 262:
These novel observations strongly s
- Page 263 and 264:
SEQUENCE DIVERGENCE IN TWO MITOCHON
- Page 265 and 266:
Table 3. Pairwise sequence distance
- Page 267 and 268:
DEVELOPMENT OF MOLECULAR DIAGNOSTIC
- Page 269 and 270:
A. B. Relative Density of SCAR Band
- Page 271 and 272:
THE ALIMENTARY TRACK OF GLASSY-WING
- Page 273 and 274:
We have dissected and identified al
- Page 275 and 276:
REALIZED LIFETIME PARASITISM AND TH
- Page 277 and 278:
REPRODUCTIVE AND DEVELOPMENTAL BIOL
- Page 279 and 280:
Mean adult longevity (days) 25 20 1
- Page 281 and 282:
the proposed exploratory work; thes
- Page 283 and 284:
SEARCHING FOR AND COLLECTING EGG PA
- Page 285 and 286:
REFERENCES Hoddle, M. S. and S. V.
- Page 287 and 288:
some Gram(+) bacteria, but are inac
- Page 289 and 290:
7. Hultmark, D., et al., Insect Imm
- Page 291 and 292:
Isolation of Fungal Pathogens Soil
- Page 293 and 294:
IDENTIFICATION OF MECHANISMS MEDIAT
- Page 295 and 296:
concentrations of 1.5 x 10 7 bacter
- Page 297 and 298:
anchor it in the outer membrane (sh
- Page 299 and 300:
SYMBIOTIC CONTROL OF PIERCE’S DIS
- Page 301 and 302:
MANAGEMENT OF PIERCE’S DISEASE OF
- Page 303 and 304:
pfF mutants are taken up by insects
- Page 305 and 306:
Simpson, A. J. G., F. C. Reinach, P
- Page 307 and 308:
Al-Wahaibi, A.K., Owen, and J. G. M
- Page 309 and 310:
patterns differed among the lines,
- Page 311 and 312:
Punja, Z.K. 2001. Genetic engineeri
- Page 313 and 314:
mind, we conducted an additional st
- Page 315 and 316:
REFERENCES Toscano, N., Byrne, F.,
- Page 317 and 318:
RESULTS AND CONCLUSIONS The program
- Page 319 and 320:
COMPATIBILITY OF INSECTICIDES WITH
- Page 321 and 322:
More tests are in progress to addre