Mechanisms of Olfaction in Insects - ResearchSpace@Auckland ...
Mechanisms of Olfaction in Insects - ResearchSpace@Auckland ...
Mechanisms of Olfaction in Insects - ResearchSpace@Auckland ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Identification <strong>of</strong> putative odorant receptors from Epiphyas postvittana 104<br />
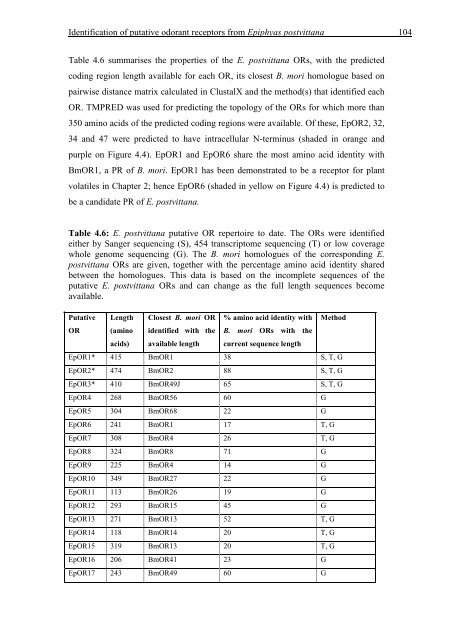
Table 4.6 summarises the properties <strong>of</strong> the E. postvittana ORs, with the predicted<br />
cod<strong>in</strong>g region length available for each OR, its closest B. mori homologue based on<br />
pairwise distance matrix calculated <strong>in</strong> ClustalX and the method(s) that identified each<br />
OR. TMPRED was used for predict<strong>in</strong>g the topology <strong>of</strong> the ORs for which more than<br />
350 am<strong>in</strong>o acids <strong>of</strong> the predicted cod<strong>in</strong>g regions were available. Of these, EpOR2, 32,<br />
34 and 47 were predicted to have <strong>in</strong>tracellular N-term<strong>in</strong>us (shaded <strong>in</strong> orange and<br />
purple on Figure 4.4). EpOR1 and EpOR6 share the most am<strong>in</strong>o acid identity with<br />
BmOR1, a PR <strong>of</strong> B. mori. EpOR1 has been demonstrated to be a receptor for plant<br />
volatiles <strong>in</strong> Chapter 2; hence EpOR6 (shaded <strong>in</strong> yellow on Figure 4.4) is predicted to<br />
be a candidate PR <strong>of</strong> E. postvittana.<br />
Table 4.6: E. postvittana putative OR repertoire to date. The ORs were identified<br />
either by Sanger sequenc<strong>in</strong>g (S), 454 transcriptome sequenc<strong>in</strong>g (T) or low coverage<br />
whole genome sequenc<strong>in</strong>g (G). The B. mori homologues <strong>of</strong> the correspond<strong>in</strong>g E.<br />
postvittana ORs are given, together with the percentage am<strong>in</strong>o acid identity shared<br />
between the homologues. This data is based on the <strong>in</strong>complete sequences <strong>of</strong> the<br />
putative E. postvittana ORs and can change as the full length sequences become<br />
available.<br />
Putative<br />
OR<br />
Length<br />
(am<strong>in</strong>o<br />
acids)<br />
Closest B. mori OR<br />
identified with the<br />
available length<br />
% am<strong>in</strong>o acid identity with<br />
B. mori ORs with the<br />
current sequence length<br />
Method<br />
EpOR1* 415 BmOR1 38 S, T, G<br />
EpOR2* 474 BmOR2 88 S, T, G<br />
EpOR3* 410 BmOR49J 65 S, T, G<br />
EpOR4 268 BmOR56 60 G<br />
EpOR5 304 BmOR68 22 G<br />
EpOR6 241 BmOR1 17 T, G<br />
EpOR7 308 BmOR4 26 T, G<br />
EpOR8 324 BmOR8 71 G<br />
EpOR9 225 BmOR4 14 G<br />
EpOR10 349 BmOR27 22 G<br />
EpOR11 113 BmOR26 19 G<br />
EpOR12 293 BmOR15 45 G<br />
EpOR13 271 BmOR13 52 T, G<br />
EpOR14 118 BmOR14 20 T, G<br />
EpOR15 319 BmOR13 20 T, G<br />
EpOR16 206 BmOR41 23 G<br />
EpOR17 243 BmOR49 60 G