in vitro PHARMACOLOGY 2011 CATALOG - Cerep
in vitro PHARMACOLOGY 2011 CATALOG - Cerep
in vitro PHARMACOLOGY 2011 CATALOG - Cerep
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
163<br />
<strong>Cerep</strong><br />
services<br />
Other enzymes<br />
Receptors<br />
Ion<br />
channels<br />
❚ Phosphatases<br />
Transporters<br />
Reversibility of prote<strong>in</strong> tyros<strong>in</strong>e residue phosphorylation is an important factor <strong>in</strong> the <strong>in</strong>tra-cellular signall<strong>in</strong>g as phosphorylation <strong>in</strong>duces<br />
conformational changes, creates dock<strong>in</strong>g sites for other prote<strong>in</strong>s and causes <strong>in</strong>tracellular relocation. This reversibility relies on the co-ord<strong>in</strong>ated<br />
actions of prote<strong>in</strong> tyros<strong>in</strong>e k<strong>in</strong>ases and prote<strong>in</strong> tyros<strong>in</strong>e phosphatases (PTPs).<br />
Prote<strong>in</strong> tyros<strong>in</strong>e phosphatases are divided <strong>in</strong> receptor PTPs (rPTPs) and non receptor PTPs (nrPTPs), the dual specificity PTP (dsPTPs) and the<br />
low molecular PTP families. They are <strong>in</strong>volved <strong>in</strong> cell-substrate adhesion, cell-cell adhesion and <strong>in</strong>sul<strong>in</strong> signall<strong>in</strong>g as described <strong>in</strong> Prote<strong>in</strong><br />
tyros<strong>in</strong>e phospshatase end signal<strong>in</strong>g, Stoker, A.S. (2005), J. Endocr<strong>in</strong>ol., 185: 19-33.<br />
In this context, <strong>Cerep</strong> has designed a high throughput profil<strong>in</strong>g platform to determ<strong>in</strong>e the <strong>in</strong>hibitory activity of compounds on the phosphates<br />
superfamily. The robustness, reproducibility and relevance of this platform were determ<strong>in</strong>ed by screen<strong>in</strong>g a panel of commercially available<br />
reference <strong>in</strong>hibitors <strong>in</strong> three <strong>in</strong>dependent experiments.<br />
K<strong>in</strong>ases<br />
Epigenetic &<br />
DNA-related<br />
enzymes<br />
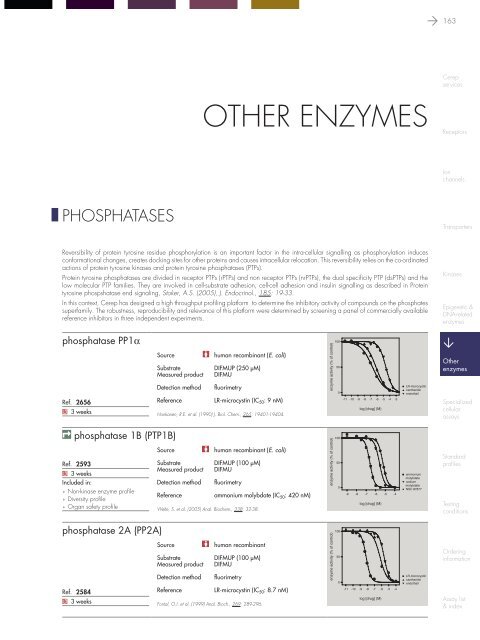
phosphatase PP1a<br />
Ref. 2656<br />
Q 3 weeks<br />
Source<br />
human recomb<strong>in</strong>ant (E. coli)<br />
Substrate<br />
DIFMUP (250 µM)<br />
Measured product DIFMU<br />
Detection method fluorimetry<br />
Reference<br />
LR-microcyst<strong>in</strong> (IC 50 : 9 nM)<br />
Honkanen, R.E. et al. (1990) J. Biol. Chem., 265: 19401-19404.<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
Other<br />
enzymes<br />
Specialized<br />
cellular<br />
assays<br />
phosphatase 1B (PTP1B)<br />
Ref. 2593<br />
Q 3 weeks<br />
Included <strong>in</strong>:<br />
Non-k<strong>in</strong>ase enzyme profile<br />
Diversity profile<br />
Organ safety profile<br />
Source<br />
Substrate<br />
Measured product<br />
Detection method<br />
Reference<br />
human recomb<strong>in</strong>ant (E. coli)<br />
DIFMUP (100 µM)<br />
DIFMU<br />
fluorimetry<br />
Welte, S. et al. (2005) Anal. Biochem., 338: 32-38.<br />
ammonium molybdate (IC 50 : 420 nM)<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
Standard<br />
profiles<br />
Test<strong>in</strong>g<br />
conditions<br />
phosphatase 2A (PP2A)<br />
Source<br />
human recomb<strong>in</strong>ant<br />
Substrate<br />
DIFMUP (100 µM)<br />
Measured product DIFMU<br />
Detection method fluorimetry<br />
Ref. 2584<br />
Reference<br />
LR-microcyst<strong>in</strong> (IC 50 : 8.7 nM)<br />
Q 3 weeks<br />
Fontal, O.I. et al. (1999) Anal. Bioch., 269: 289-296.<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
Order<strong>in</strong>g<br />
<strong>in</strong>formation<br />
Assay list<br />
& <strong>in</strong>dex