in vitro PHARMACOLOGY 2011 CATALOG - Cerep
in vitro PHARMACOLOGY 2011 CATALOG - Cerep
in vitro PHARMACOLOGY 2011 CATALOG - Cerep
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
5<br />
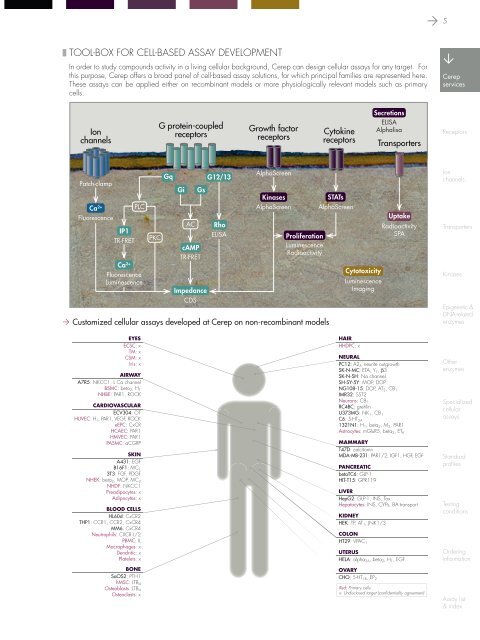
❚ tool-box for cell-based assay development<br />
In order to study compounds activity <strong>in</strong> a liv<strong>in</strong>g cellular background, <strong>Cerep</strong> can design cellular assays for any target. For<br />
this purpose, <strong>Cerep</strong> offers a broad panel of cell-based assay solutions, for which pr<strong>in</strong>cipal families are represented here.<br />
These assays can be applied either on recomb<strong>in</strong>ant models or more physiologically relevant models such as primary<br />
cells.<br />
<br />
<strong>Cerep</strong><br />
services<br />
Ion<br />
channels<br />
G prote<strong>in</strong>-coupled<br />
receptors<br />
Growth factor<br />
receptors<br />
Cytok<strong>in</strong>e<br />
receptors<br />
Secretions<br />
ELISA<br />
Alphalisa<br />
Transporters<br />
Receptors<br />
Patch-clamp<br />
Ca 2+<br />
Fluorescence<br />
IP1<br />
TR-FRET<br />
Ca 2+<br />
PLC<br />
Fluorescence<br />
Lum<strong>in</strong>escence<br />
PKC<br />
Gq<br />
Gi<br />
AC<br />
cAMP<br />
TR-FRET<br />
Impedance<br />
CDS<br />
Gs<br />
G12/13<br />
Customized cellular assays developed at <strong>Cerep</strong> on non-recomb<strong>in</strong>ant models<br />
Rho<br />
ELISA<br />
AlphaScreen<br />
K<strong>in</strong>ases<br />
AlphaScreen<br />
Proliferation<br />
Lum<strong>in</strong>escence<br />
Radioactivity<br />
STATs<br />
AlphaScreen<br />
Cytotoxicity<br />
Lum<strong>in</strong>escence<br />
Imag<strong>in</strong>g<br />
Uptake<br />
Radioactivity<br />
SPA<br />
Ion<br />
channels<br />
Transporters<br />
K<strong>in</strong>ases<br />
Epigenetic &<br />
DNA-related<br />
enzymes<br />
EYES<br />
ECSC: x<br />
TM: x<br />
CSM: x<br />
Iris: x<br />
AIRWAY<br />
A7R5: NKCC1, L Ca channel<br />
BSMC: beta 2 , H 1<br />
NHBE: PAR1, ROCK<br />
CARDIOVASCULAR<br />
ECV304: OT<br />
HUVEC: H 1 , PAR1, VEGF, ROCK<br />
eEPC: CxCR<br />
HCAEC: PAR1<br />
HMVEC: PAR1<br />
PASMC: aCGRP<br />
SKIN<br />
A431: EGF<br />
B16F1: MC 1<br />
3T3: FGF, PDGF<br />
NHEK: beta 2 , MOP, MC 2<br />
NHDF: NKCC1<br />
Preadipocytes: x<br />
Adipocytes: x<br />
BLOOD CELLS<br />
HL60d: CxCR2<br />
THP1: CCR1, CCR2, CxCR4<br />
MM6: CxCR4<br />
Neutrophils: CXCR1/2<br />
PBMC: IL<br />
Macrophages: x<br />
Dendritic: x<br />
Platelets: x<br />
BONE<br />
SaOS2: PTH1<br />
hMSC: LTB 4<br />
Osteoblasts: LTB 4<br />
Osteoclasts: x<br />
HAIR<br />
HHDPC: x<br />
NEURAL<br />
PC12: A2 A, neurite outgrowth<br />
SK-N-MC: ETA, Y 1 , β3<br />
SK-N-SH: Na channel<br />
SH-SY-5Y: MOP, DOP<br />
NG108-15: DOP, AT 2 , CB 1<br />
IMR32: SST2<br />
Neurons: CB 1<br />
RC4BC: grehl<strong>in</strong><br />
U373MG: NK 1 , CB 1<br />
C6: 5-HT 2A<br />
1321N1: H 1 , beta 2 , M 3 , PAR1<br />
Astrocytes: mGluR5, beta 2 , ET A<br />
MAMMARY<br />
T47D: calciton<strong>in</strong><br />
MDA-MB-231: PAR1/2, IGF1, HGF, EGF<br />
PANCREATIC<br />
betaTC6: GLP-1<br />
HIT-T15: GPR119<br />
LIVER<br />
HepG2: GLP-1, INS, Tox<br />
Hepatocytes: INS, CYPs, BA transport<br />
KIDNEY<br />
HEK: TP, AT 1 , JNK1/3<br />
COLON<br />
HT29: VPAC 1<br />
UTERUS<br />
HELA: alpha 2A , beta 2 , H 1 , EGF<br />
OVARY<br />
CHO: 5-HT 1B , EP 2<br />
Red: Primary cells<br />
x: Undisclosed target (confi dentiality agreement)<br />
Other<br />
enzymes<br />
Specialized<br />
cellular<br />
assays<br />
Standard<br />
profiles<br />
Test<strong>in</strong>g<br />
conditions<br />
Order<strong>in</strong>g<br />
<strong>in</strong>formation<br />
Assay list<br />
& <strong>in</strong>dex