The Genom of Homo sapiens.pdf
The Genom of Homo sapiens.pdf
The Genom of Homo sapiens.pdf
- TAGS
- homo
- www.yumpu.com
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
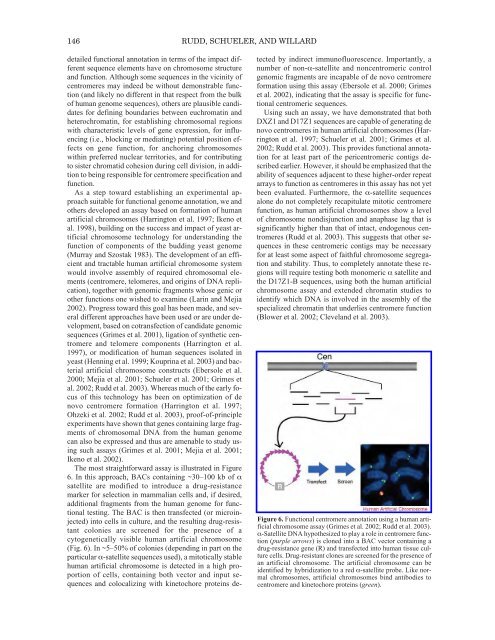
146 RUDD, SCHUELER, AND WILLARDdetailed functional annotation in terms <strong>of</strong> the impact differentsequence elements have on chromosome structureand function. Although some sequences in the vicinity <strong>of</strong>centromeres may indeed be without demonstrable function(and likely no different in that respect from the bulk<strong>of</strong> human genome sequences), others are plausible candidatesfor defining boundaries between euchromatin andheterochromatin, for establishing chromosomal regionswith characteristic levels <strong>of</strong> gene expression, for influencing(i.e., blocking or mediating) potential position effectson gene function, for anchoring chromosomeswithin preferred nuclear territories, and for contributingto sister chromatid cohesion during cell division, in additionto being responsible for centromere specification andfunction.As a step toward establishing an experimental approachsuitable for functional genome annotation, we andothers developed an assay based on formation <strong>of</strong> humanartificial chromosomes (Harrington et al. 1997; Ikeno etal. 1998), building on the success and impact <strong>of</strong> yeast artificialchromosome technology for understanding thefunction <strong>of</strong> components <strong>of</strong> the budding yeast genome(Murray and Szostak 1983). <strong>The</strong> development <strong>of</strong> an efficientand tractable human artificial chromosome systemwould involve assembly <strong>of</strong> required chromosomal elements(centromere, telomeres, and origins <strong>of</strong> DNA replication),together with genomic fragments whose genic orother functions one wished to examine (Larin and Mejia2002). Progress toward this goal has been made, and severaldifferent approaches have been used or are under development,based on cotransfection <strong>of</strong> candidate genomicsequences (Grimes et al. 2001), ligation <strong>of</strong> synthetic centromereand telomere components (Harrington et al.1997), or modification <strong>of</strong> human sequences isolated inyeast (Henning et al. 1999; Kouprina et al. 2003) and bacterialartificial chromosome constructs (Ebersole et al.2000; Mejia et al. 2001; Schueler et al. 2001; Grimes etal. 2002; Rudd et al. 2003). Whereas much <strong>of</strong> the early focus<strong>of</strong> this technology has been on optimization <strong>of</strong> denovo centromere formation (Harrington et al. 1997;Ohzeki et al. 2002; Rudd et al. 2003), pro<strong>of</strong>-<strong>of</strong>-principleexperiments have shown that genes containing large fragments<strong>of</strong> chromosomal DNA from the human genomecan also be expressed and thus are amenable to study usingsuch assays (Grimes et al. 2001; Mejia et al. 2001;Ikeno et al. 2002).<strong>The</strong> most straightforward assay is illustrated in Figure6. In this approach, BACs containing ~30–100 kb <strong>of</strong> αsatellite are modified to introduce a drug-resistancemarker for selection in mammalian cells and, if desired,additional fragments from the human genome for functionaltesting. <strong>The</strong> BAC is then transfected (or microinjected)into cells in culture, and the resulting drug-resistantcolonies are screened for the presence <strong>of</strong> acytogenetically visible human artificial chromosome(Fig. 6). In ~5–50% <strong>of</strong> colonies (depending in part on theparticular α-satellite sequences used), a mitotically stablehuman artificial chromosome is detected in a high proportion<strong>of</strong> cells, containing both vector and input sequencesand colocalizing with kinetochore proteins detectedby indirect immun<strong>of</strong>luorescence. Importantly, anumber <strong>of</strong> non-α-satellite and noncentromeric controlgenomic fragments are incapable <strong>of</strong> de novo centromereformation using this assay (Ebersole et al. 2000; Grimeset al. 2002), indicating that the assay is specific for functionalcentromeric sequences.Using such an assay, we have demonstrated that bothDXZ1 and D17Z1 sequences are capable <strong>of</strong> generating denovo centromeres in human artificial chromosomes (Harringtonet al. 1997; Schueler et al. 2001; Grimes et al.2002; Rudd et al. 2003). This provides functional annotationfor at least part <strong>of</strong> the pericentromeric contigs describedearlier. However, it should be emphasized that theability <strong>of</strong> sequences adjacent to these higher-order repeatarrays to function as centromeres in this assay has not yetbeen evaluated. Furthermore, the α-satellite sequencesalone do not completely recapitulate mitotic centromerefunction, as human artificial chromosomes show a level<strong>of</strong> chromosome nondisjunction and anaphase lag that issignificantly higher than that <strong>of</strong> intact, endogenous centromeres(Rudd et al. 2003). This suggests that other sequencesin these centromeric contigs may be necessaryfor at least some aspect <strong>of</strong> faithful chromosome segregationand stability. Thus, to completely annotate these regionswill require testing both monomeric α satellite andthe D17Z1-B sequences, using both the human artificialchromosome assay and extended chromatin studies toidentify which DNA is involved in the assembly <strong>of</strong> thespecialized chromatin that underlies centromere function(Blower et al. 2002; Cleveland et al. 2003).Figure 6. Functional centromere annotation using a human artificialchromosome assay (Grimes et al. 2002; Rudd et al. 2003).α-Satellite DNA hypothesized to play a role in centromere function(purple arrows) is cloned into a BAC vector containing adrug-resistance gene (R) and transfected into human tissue culturecells. Drug-resistant clones are screened for the presence <strong>of</strong>an artificial chromosome. <strong>The</strong> artificial chromosome can beidentified by hybridization to a red α-satellite probe. Like normalchromosomes, artificial chromosomes bind antibodies tocentromere and kinetochore proteins (green).