The Genom of Homo sapiens.pdf
The Genom of Homo sapiens.pdf
The Genom of Homo sapiens.pdf
- TAGS
- homo
- www.yumpu.com
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
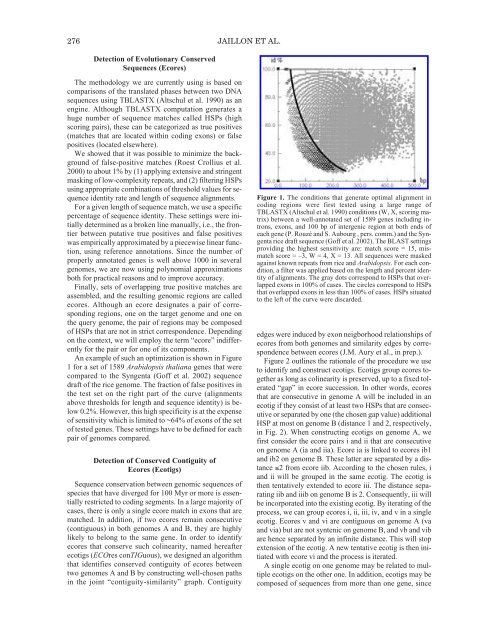
276 JAILLON ET AL.Detection <strong>of</strong> Evolutionary ConservedSequences (Ecores)<strong>The</strong> methodology we are currently using is based oncomparisons <strong>of</strong> the translated phases between two DNAsequences using TBLASTX (Altschul et al. 1990) as anengine. Although TBLASTX computation generates ahuge number <strong>of</strong> sequence matches called HSPs (highscoring pairs), these can be categorized as true positives(matches that are located within coding exons) or falsepositives (located elsewhere).We showed that it was possible to minimize the background<strong>of</strong> false-positive matches (Roest Crollius et al.2000) to about 1% by (1) applying extensive and stringentmasking <strong>of</strong> low-complexity repeats, and (2) filtering HSPsusing appropriate combinations <strong>of</strong> threshold values for sequenceidentity rate and length <strong>of</strong> sequence alignments.For a given length <strong>of</strong> sequence match, we use a specificpercentage <strong>of</strong> sequence identity. <strong>The</strong>se settings were initiallydetermined as a broken line manually, i.e., the frontierbetween putative true positives and false positiveswas empirically approximated by a piecewise linear function,using reference annotations. Since the number <strong>of</strong>properly annotated genes is well above 1000 in severalgenomes, we are now using polynomial approximationsboth for practical reasons and to improve accuracy.Finally, sets <strong>of</strong> overlapping true positive matches areassembled, and the resulting genomic regions are calledecores. Although an ecore designates a pair <strong>of</strong> correspondingregions, one on the target genome and one onthe query genome, the pair <strong>of</strong> regions may be composed<strong>of</strong> HSPs that are not in strict correspondence. Dependingon the context, we will employ the term “ecore” indifferentlyfor the pair or for one <strong>of</strong> its components.An example <strong>of</strong> such an optimization is shown in Figure1 for a set <strong>of</strong> 1589 Arabidopsis thaliana genes that werecompared to the Syngenta (G<strong>of</strong>f et al. 2002) sequencedraft <strong>of</strong> the rice genome. <strong>The</strong> fraction <strong>of</strong> false positives inthe test set on the right part <strong>of</strong> the curve (alignmentsabove thresholds for length and sequence identity) is below0.2%. However, this high specificity is at the expense<strong>of</strong> sensitivity which is limited to ~64% <strong>of</strong> exons <strong>of</strong> the set<strong>of</strong> tested genes. <strong>The</strong>se settings have to be defined for eachpair <strong>of</strong> genomes compared.Detection <strong>of</strong> Conserved Contiguity <strong>of</strong>Ecores (Ecotigs)Sequence conservation between genomic sequences <strong>of</strong>species that have diverged for 100 Myr or more is essentiallyrestricted to coding segments. In a large majority <strong>of</strong>cases, there is only a single ecore match in exons that arematched. In addition, if two ecores remain consecutive(contiguous) in both genomes A and B, they are highlylikely to belong to the same gene. In order to identifyecores that conserve such colinearity, named hereafterecotigs (ECOres conTIGuous), we designed an algorithmthat identifies conserved contiguity <strong>of</strong> ecores betweentwo genomes A and B by constructing well-chosen pathsin the joint “contiguity-similarity” graph. ContiguityFigure 1. <strong>The</strong> conditions that generate optimal alignment incoding regions were first tested using a large range <strong>of</strong>TBLASTX (Altschul et al. 1990) conditions (W, X, scoring matrix)between a well-annotated set <strong>of</strong> 1589 genes including introns,exons, and 100 bp <strong>of</strong> intergenic region at both ends <strong>of</strong>each gene (P. Rouzé and S. Aubourg , pers. comm.) and the Syngentarice draft sequence (G<strong>of</strong>f et al. 2002). <strong>The</strong> BLAST settingsproviding the highest sensitivity are: match score = 15, mismatchscore = –3, W = 4, X = 13. All sequences were maskedagainst known repeats from rice and Arabidopsis. For each condition,a filter was applied based on the length and percent identity<strong>of</strong> alignments. <strong>The</strong> gray dots correspond to HSPs that overlappedexons in 100% <strong>of</strong> cases. <strong>The</strong> circles correspond to HSPsthat overlapped exons in less than 100% <strong>of</strong> cases. HSPs situatedto the left <strong>of</strong> the curve were discarded.edges were induced by exon neigborhood relationships <strong>of</strong>ecores from both genomes and similarity edges by correspondencebetween ecores (J.M. Aury et al., in prep.).Figure 2 outlines the rationale <strong>of</strong> the procedure we useto identify and construct ecotigs. Ecotigs group ecores togetheras long as colinearity is preserved, up to a fixed tolerated“gap” in ecore succession. In other words, ecoresthat are consecutive in genome A will be included in anecotig if they consist <strong>of</strong> at least two HSPs that are consecutiveor separated by one (the chosen gap value) additionalHSP at most on genome B (distance 1 and 2, respectively,in Fig. 2). When constructing ecotigs on genome A, wefirst consider the ecore pairs i and ii that are consecutiveon genome A (ia and iia). Ecore ia is linked to ecores ib1and ib2 on genome B. <strong>The</strong>se latter are separated by a distance≤2 from ecore iib. According to the chosen rules, iand ii will be grouped in the same ecotig. <strong>The</strong> ecotig isthen tentatively extended to ecore iii. <strong>The</strong> distance separatingiib and iiib on genome B is 2. Consequently, iii willbe incorporated into the existing ecotig. By iterating <strong>of</strong> theprocess, we can group ecores i, ii, iii, iv, and v in a singleecotig. Ecores v and vi are contiguous on genome A (vaand via) but are not syntenic on genome B, and vb and vibare hence separated by an infinite distance. This will stopextension <strong>of</strong> the ecotig. A new tentative ecotig is then initiatedwith ecore vi and the process is iterated.A single ecotig on one genome may be related to multipleecotigs on the other one. In addition, ecotigs may becomposed <strong>of</strong> sequences from more than one gene, since