The Genom of Homo sapiens.pdf
The Genom of Homo sapiens.pdf
The Genom of Homo sapiens.pdf
- TAGS
- homo
- www.yumpu.com
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
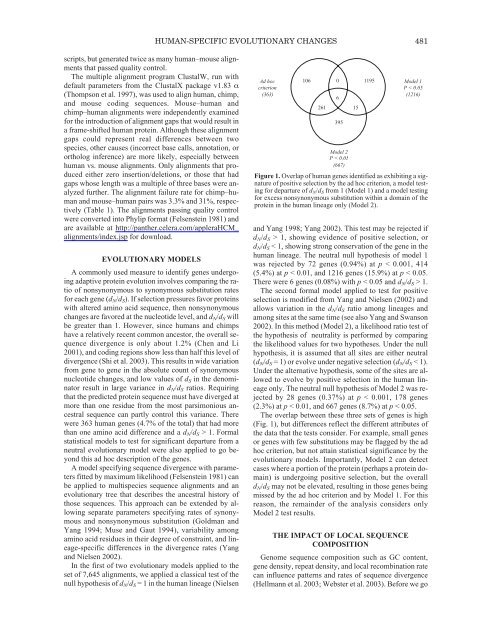
HUMAN-SPECIFIC EVOLUTIONARY CHANGES 481scripts, but generated twice as many human–mouse alignmentsthat passed quality control.<strong>The</strong> multiple alignment program ClustalW, run withdefault parameters from the ClustalX package v1.83 α(Thompson et al. 1997), was used to align human, chimp,and mouse coding sequences. Mouse–human andchimp–human alignments were independently examinedfor the introduction <strong>of</strong> alignment gaps that would result ina frame-shifted human protein. Although these alignmentgaps could represent real differences between twospecies, other causes (incorrect base calls, annotation, orortholog inference) are more likely, especially betweenhuman vs. mouse alignments. Only alignments that producedeither zero insertion/deletions, or those that hadgaps whose length was a multiple <strong>of</strong> three bases were analyzedfurther. <strong>The</strong> alignment failure rate for chimp–humanand mouse–human pairs was 3.3% and 31%, respectively(Table 1). <strong>The</strong> alignments passing quality controlwere converted into Phylip format (Felsenstein 1981) andare available at http://panther.celera.com/appleraHCM_alignments/index.jsp for download.EVOLUTIONARY MODELSA commonly used measure to identify genes undergoingadaptive protein evolution involves comparing the ratio<strong>of</strong> nonsynonymous to synonymous substitution ratesfor each gene (d N /d S ). If selection pressures favor proteinswith altered amino acid sequence, then nonsynonymouschanges are favored at the nucleotide level, and d N /d S willbe greater than 1. However, since humans and chimpshave a relatively recent common ancestor, the overall sequencedivergence is only about 1.2% (Chen and Li2001), and coding regions show less than half this level <strong>of</strong>divergence (Shi et al. 2003). This results in wide variationfrom gene to gene in the absolute count <strong>of</strong> synonymousnucleotide changes, and low values <strong>of</strong> d S in the denominatorresult in large variance in d N /d S ratios. Requiringthat the predicted protein sequence must have diverged atmore than one residue from the most parsimonious ancestralsequence can partly control this variance. <strong>The</strong>rewere 363 human genes (4.7% <strong>of</strong> the total) that had morethan one amino acid difference and a d N /d S > 1. Formalstatistical models to test for significant departure from aneutral evolutionary model were also applied to go beyondthis ad hoc description <strong>of</strong> the genes.A model specifying sequence divergence with parametersfitted by maximum likelihood (Felsenstein 1981) canbe applied to multispecies sequence alignments and anevolutionary tree that describes the ancestral history <strong>of</strong>those sequences. This approach can be extended by allowingseparate parameters specifying rates <strong>of</strong> synonymousand nonsynonymous substitution (Goldman andYang 1994; Muse and Gaut 1994), variability amongamino acid residues in their degree <strong>of</strong> constraint, and lineage-specificdifferences in the divergence rates (Yangand Nielsen 2002).In the first <strong>of</strong> two evolutionary models applied to theset <strong>of</strong> 7,645 alignments, we applied a classical test <strong>of</strong> thenull hypothesis <strong>of</strong> d N /d S = 1 in the human lineage (NielsenAd hoccriterion(363)106 02616395and Yang 1998; Yang 2002). This test may be rejected ifd N /d S > 1, showing evidence <strong>of</strong> positive selection, ord N /d S < 1, showing strong conservation <strong>of</strong> the gene in thehuman lineage. <strong>The</strong> neutral null hypothesis <strong>of</strong> model 1was rejected by 72 genes (0.94%) at p < 0.001, 414(5.4%) at p < 0.01, and 1216 genes (15.9%) at p < 0.05.<strong>The</strong>re were 6 genes (0.08%) with p < 0.05 and d N /d S > 1.<strong>The</strong> second formal model applied to test for positiveselection is modified from Yang and Nielsen (2002) andallows variation in the d N /d S ratio among lineages andamong sites at the same time (see also Yang and Swanson2002). In this method (Model 2), a likelihood ratio test <strong>of</strong>the hypothesis <strong>of</strong> neutrality is performed by comparingthe likelihood values for two hypotheses. Under the nullhypothesis, it is assumed that all sites are either neutral(d N /d S = 1) or evolve under negative selection (d N /d S < 1).Under the alternative hypothesis, some <strong>of</strong> the sites are allowedto evolve by positive selection in the human lineageonly. <strong>The</strong> neutral null hypothesis <strong>of</strong> Model 2 was rejectedby 28 genes (0.37%) at p < 0.001, 178 genes(2.3%) at p < 0.01, and 667 genes (8.7%) at p < 0.05.<strong>The</strong> overlap between these three sets <strong>of</strong> genes is high(Fig. 1), but differences reflect the different attributes <strong>of</strong>the data that the tests consider. For example, small genesor genes with few substitutions may be flagged by the adhoc criterion, but not attain statistical significance by theevolutionary models. Importantly, Model 2 can detectcases where a portion <strong>of</strong> the protein (perhaps a protein domain)is undergoing positive selection, but the overalld N /d S may not be elevated, resulting in those genes beingmissed by the ad hoc criterion and by Model 1. For thisreason, the remainder <strong>of</strong> the analysis considers onlyModel 2 test results.THE IMPACT OF LOCAL SEQUENCECOMPOSITION<strong>Genom</strong>e sequence composition such as GC content,gene density, repeat density, and local recombination ratecan influence patterns and rates <strong>of</strong> sequence divergence(Hellmann et al. 2003; Webster et al. 2003). Before we go151195Model 1P < 0.05(1216)Model 2P < 0.05(667)Figure 1. Overlap <strong>of</strong> human genes identified as exhibiting a signature<strong>of</strong> positive selection by the ad hoc criterion, a model testingfor departure <strong>of</strong> d N /d S from 1 (Model 1) and a model testingfor excess nonsynonymous substitution within a domain <strong>of</strong> theprotein in the human lineage only (Model 2).