The Genom of Homo sapiens.pdf
The Genom of Homo sapiens.pdf
The Genom of Homo sapiens.pdf
- TAGS
- homo
- www.yumpu.com
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
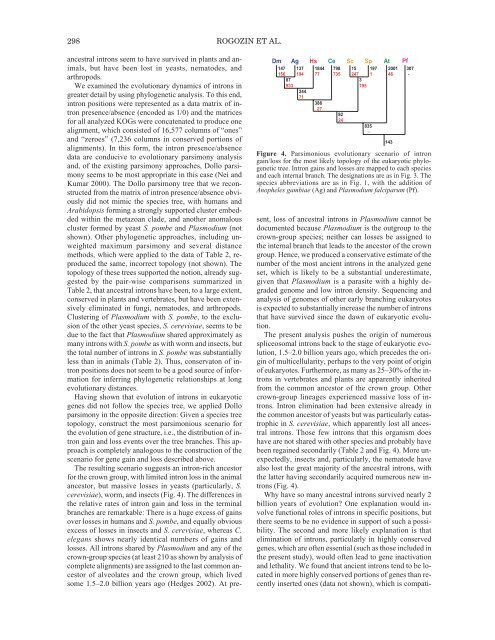
298 ROGOZIN ET AL.Dm Ag Hs Ce Sc Sp At Pf147156137194184477798735152471971200146307-87933379524471386279224835-Figure 4. Parsimonious evolutionary scenario <strong>of</strong> introngain/loss for the most likely topology <strong>of</strong> the eukaryotic phylogenetictree. Intron gains and losses are mapped to each speciesand each internal branch. <strong>The</strong> designations are as in Fig. 3. <strong>The</strong>species abbreviations are as in Fig. 1, with the addition <strong>of</strong>Anopheles gambiae (Ag) and Plasmodium falciparum (Pf).ancestral introns seem to have survived in plants and animals,but have been lost in yeasts, nematodes, andarthropods.We examined the evolutionary dynamics <strong>of</strong> introns ingreater detail by using phylogenetic analysis. To this end,intron positions were represented as a data matrix <strong>of</strong> intronpresence/absence (encoded as 1/0) and the matricesfor all analyzed KOGs were concatenated to produce onealignment, which consisted <strong>of</strong> 16,577 columns <strong>of</strong> “ones”and “zeroes” (7,236 columns in conserved portions <strong>of</strong>alignments). In this form, the intron presence/absencedata are conducive to evolutionary parsimony analysisand, <strong>of</strong> the existing parsimony approaches, Dollo parsimonyseems to be most appropriate in this case (Nei andKumar 2000). <strong>The</strong> Dollo parsimony tree that we reconstructedfrom the matrix <strong>of</strong> intron presence/absence obviouslydid not mimic the species tree, with humans andArabidopsis forming a strongly supported cluster embeddedwithin the metazoan clade, and another anomalouscluster formed by yeast S. pombe and Plasmodium (notshown). Other phylogenetic approaches, including unweightedmaximum parsimony and several distancemethods, which were applied to the data <strong>of</strong> Table 2, reproducedthe same, incorrect topology (not shown). <strong>The</strong>topology <strong>of</strong> these trees supported the notion, already suggestedby the pair-wise comparisons summarized inTable 2, that ancestral introns have been, to a large extent,conserved in plants and vertebrates, but have been extensivelyeliminated in fungi, nematodes, and arthropods.Clustering <strong>of</strong> Plasmodium with S. pombe, to the exclusion<strong>of</strong> the other yeast species, S. cerevisiae, seems to bedue to the fact that Plasmodium shared approximately asmany introns with S. pombe as with worm and insects, butthe total number <strong>of</strong> introns in S. pombe was substantiallyless than in animals (Table 2). Thus, conservaton <strong>of</strong> intronpositions does not seem to be a good source <strong>of</strong> informationfor inferring phylogenetic relationships at longevolutionary distances.Having shown that evolution <strong>of</strong> introns in eukaryoticgenes did not follow the species tree, we applied Dolloparsimony in the opposite direction: Given a species treetopology, construct the most parsimonious scenario forthe evolution <strong>of</strong> gene structure, i.e., the distribution <strong>of</strong> introngain and loss events over the tree branches. This approachis completely analogous to the construction <strong>of</strong> thescenario for gene gain and loss described above.<strong>The</strong> resulting scenario suggests an intron-rich ancestorfor the crown group, with limited intron loss in the animalancestor, but massive losses in yeasts (particularly, S.cerevisiae), worm, and insects (Fig. 4). <strong>The</strong> differences inthe relative rates <strong>of</strong> intron gain and loss in the terminalbranches are remarkable: <strong>The</strong>re is a huge excess <strong>of</strong> gainsover losses in humans and S. pombe, and equally obviousexcess <strong>of</strong> losses in insects and S. cerevisiae, whereas C.elegans shows nearly identical numbers <strong>of</strong> gains andlosses. All introns shared by Plasmodium and any <strong>of</strong> thecrown-group species (at least 210 as shown by analysis <strong>of</strong>complete alignments) are assigned to the last common ancestor<strong>of</strong> alveolates and the crown group, which livedsome 1.5–2.0 billion years ago (Hedges 2002). At present,loss <strong>of</strong> ancestral introns in Plasmodium cannot bedocumented because Plasmodium is the outgroup to thecrown-group species; neither can losses be assigned tothe internal branch that leads to the ancestor <strong>of</strong> the crowngroup. Hence, we produced a conservative estimate <strong>of</strong> thenumber <strong>of</strong> the most ancient introns in the analyzed geneset, which is likely to be a substantial underestimate,given that Plasmodium is a parasite with a highly degradedgenome and low intron density. Sequencing andanalysis <strong>of</strong> genomes <strong>of</strong> other early branching eukaryotesis expected to substantially increase the number <strong>of</strong> intronsthat have survived since the dawn <strong>of</strong> eukaryotic evolution.<strong>The</strong> present analysis pushes the origin <strong>of</strong> numerousspliceosomal introns back to the stage <strong>of</strong> eukaryotic evolution,1.5–2.0 billion years ago, which precedes the origin<strong>of</strong> multicellularity, perhaps to the very point <strong>of</strong> origin<strong>of</strong> eukaryotes. Furthermore, as many as 25–30% <strong>of</strong> the intronsin vertebrates and plants are apparently inheritedfrom the common ancestor <strong>of</strong> the crown group. Othercrown-group lineages experienced massive loss <strong>of</strong> introns.Intron elimination had been extensive already inthe common ancestor <strong>of</strong> yeasts but was particularly catastrophicin S. cerevisiae, which apparently lost all ancestralintrons. Those few introns that this organism doeshave are not shared with other species and probably havebeen regained secondarily (Table 2 and Fig. 4). More unexpectedly,insects and, particularly, the nematode havealso lost the great majority <strong>of</strong> the ancestral introns, withthe latter having secondarily acquired numerous new introns(Fig. 4).Why have so many ancestral introns survived nearly 2billion years <strong>of</strong> evolution? One explanation would involvefunctional roles <strong>of</strong> introns in specific positions, butthere seems to be no evidence in support <strong>of</strong> such a possibility.<strong>The</strong> second and more likely explanation is thatelimination <strong>of</strong> introns, particularly in highly conservedgenes, which are <strong>of</strong>ten essential (such as those included inthe present study), would <strong>of</strong>ten lead to gene inactivationand lethality. We found that ancient introns tend to be locatedin more highly conserved portions <strong>of</strong> genes than recentlyinserted ones (data not shown), which is compati-143