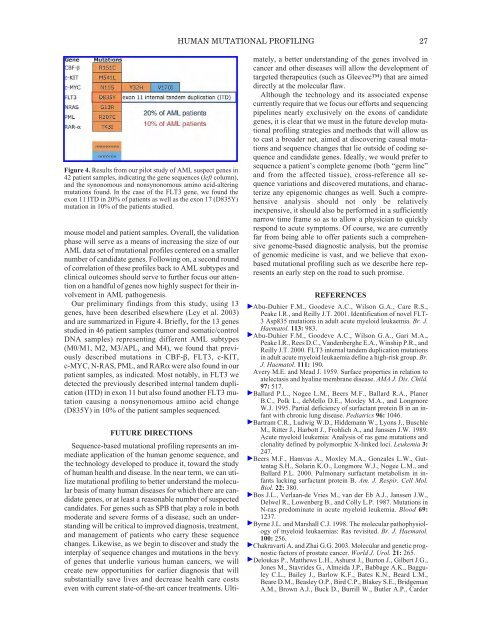
HUMAN MUTATIONAL PROFILING 27Figure 4. Results from our pilot study <strong>of</strong> AML suspect genes in42 patient samples, indicating the gene sequences (left column),and the synonomous and nonsynonomous amino acid-alteringmutations found. In the case <strong>of</strong> the FLT3 gene, we found theexon 11 ITD in 20% <strong>of</strong> patients as well as the exon 17 (D835Y)mutation in 10% <strong>of</strong> the patients studied.mouse model and patient samples. Overall, the validationphase will serve as a means <strong>of</strong> increasing the size <strong>of</strong> ourAML data set <strong>of</strong> mutational pr<strong>of</strong>iles centered on a smallernumber <strong>of</strong> candidate genes. Following on, a second round<strong>of</strong> correlation <strong>of</strong> these pr<strong>of</strong>iles back to AML subtypes andclinical outcomes should serve to further focus our attentionon a handful <strong>of</strong> genes now highly suspect for their involvementin AML pathogenesis.Our preliminary findings from this study, using 13genes, have been described elsewhere (Ley et al. 2003)and are summarized in Figure 4. Briefly, for the 13 genesstudied in 46 patient samples (tumor and somatic/controlDNA samples) representing different AML subtypes(M0/M1, M2, M3/APL, and M4), we found that previouslydescribed mutations in CBF-β, FLT3, c-KIT,c-MYC, N-RAS, PML, and RARα were also found in ourpatient samples, as indicated. Most notably, in FLT3 wedetected the previously described internal tandem duplication(ITD) in exon 11 but also found another FLT3 mutationcausing a nonsynonomous amino acid change(D835Y) in 10% <strong>of</strong> the patient samples sequenced.FUTURE DIRECTIONSSequence-based mutational pr<strong>of</strong>iling represents an immediateapplication <strong>of</strong> the human genome sequence, andthe technology developed to produce it, toward the study<strong>of</strong> human health and disease. In the near term, we can utilizemutational pr<strong>of</strong>iling to better understand the molecularbasis <strong>of</strong> many human diseases for which there are candidategenes, or at least a reasonable number <strong>of</strong> suspectedcandidates. For genes such as SPB that play a role in bothmoderate and severe forms <strong>of</strong> a disease, such an understandingwill be critical to improved diagnosis, treatment,and management <strong>of</strong> patients who carry these sequencechanges. Likewise, as we begin to discover and study theinterplay <strong>of</strong> sequence changes and mutations in the bevy<strong>of</strong> genes that underlie various human cancers, we willcreate new opportunities for earlier diagnosis that willsubstantially save lives and decrease health care costseven with current state-<strong>of</strong>-the-art cancer treatments. Ultimately,a better understanding <strong>of</strong> the genes involved incancer and other diseases will allow the development <strong>of</strong>targeted therapeutics (such as Gleevec) that are aimeddirectly at the molecular flaw.Although the technology and its associated expensecurrently require that we focus our efforts and sequencingpipelines nearly exclusively on the exons <strong>of</strong> candidategenes, it is clear that we must in the future develop mutationalpr<strong>of</strong>iling strategies and methods that will allow usto cast a broader net, aimed at discovering causal mutationsand sequence changes that lie outside <strong>of</strong> coding sequenceand candidate genes. Ideally, we would prefer tosequence a patient’s complete genome (both “germ line”and from the affected tissue), cross-reference all sequencevariations and discovered mutations, and characterizeany epigenomic changes as well. Such a comprehensiveanalysis should not only be relativelyinexpensive, it should also be performed in a sufficientlynarrow time frame so as to allow a physician to quicklyrespond to acute symptoms. Of course, we are currentlyfar from being able to <strong>of</strong>fer patients such a comprehensivegenome-based diagnostic analysis, but the promise<strong>of</strong> genomic medicine is vast, and we believe that exonbasedmutational pr<strong>of</strong>iling such as we describe here representsan early step on the road to such promise.REFERENCESAbu-Duhier F.M., Goodeve A.C., Wilson G.A., Care R.S.,Peake I.R., and Reilly J.T. 2001. Identification <strong>of</strong> novel FLT-3 Asp835 mutations in adult acute myeloid leukaemia. Br. J.Haematol. 113: 983.Abu-Duhier F.M., Goodeve A.C., Wilson G.A., Gari M.A.,Peake I.R., Rees D.C., Vandenberghe E.A., Winship P.R., andReilly J.T. 2000. FLT3 internal tandem duplication mutationsin adult acute myeloid leukaemia define a high-risk group. Br.J. Haematol. 111: 190.Avery M.E. and Mead J. 1959. Surface properties in relation toatelectasis and hyaline membrane disease. AMA J. Dis. Child.97: 517.Ballard P.L., Nogee L.M., Beers M.F., Ballard R.A., PlanerB.C., Polk L., deMello D.E., Moxley M.A., and LongmoreW.J. 1995. Partial deficiency <strong>of</strong> surfactant protein B in an infantwith chronic lung disease. Pediatrics 96: 1046.Bartram C.R., Ludwig W.D., Hiddemann W., Lyons J., BuschleM., Ritter J., Harbott J., Frohlich A., and Janssen J.W. 1989.Acute myeloid leukemia: Analysis <strong>of</strong> ras gene mutations andclonality defined by polymorphic X-linked loci. Leukemia 3:247.Beers M.F., Hamvas A., Moxley M.A., Gonzales L.W., GuttentagS.H., Solarin K.O., Longmore W.J., Nogee L.M., andBallard P.L. 2000. Pulmonary surfactant metabolism in infantslacking surfactant protein B. Am. J. Respir. Cell Mol.Biol. 22: 380.Bos J.L., Verlaan-de Vries M., van der Eb A.J., Janssen J.W.,Delwel R., Lowenberg B., and Colly L.P. 1987. Mutations inN-ras predominate in acute myeloid leukemia. Blood 69:1237.Byrne J.L. and Marshall C.J. 1998. <strong>The</strong> molecular pathophysiology<strong>of</strong> myeloid leukaemias: Ras revisited. Br. J. Haematol.100: 256.Chakravarti A. and Zhai G.G. 2003. Molecular and genetic prognosticfactors <strong>of</strong> prostate cancer. World J. Urol. 21: 265.Deloukas P., Matthews L.H., Ashurst J., Burton J., Gilbert J.G.,Jones M., Stavrides G., Almeida J.P., Babbage A.K., BagguleyC.L., Bailey J., Barlow K.F., Bates K.N., Beard L.M.,Beare D.M., Beasley O.P., Bird C.P., Blakey S.E., BridgemanA.M., Brown A.J., Buck D., Burrill W., Butler A.P., Carder
28 WILSON ET AL.C., and Carter N.P., et al. 2001. <strong>The</strong> DNA sequence and comparativeanalysis <strong>of</strong> human chromosome 20. Nature 414: 865.Dunham I., Shimizu N., Roe .A., Chissoe S., Hunt A.R., CollinsJ.E., Bruskiewich R., Beare D.M., Clamp M., Smink L.J.,Ainscough R., Almeida J.P., Babbage A., Bagguley C., BaileyJ., Barlow K., Bates K.N., Beasley O., Bird C.P., BlakeyS., Bridgeman A.M., Buck D., Burgess J., Burrill W.D., andO’Brien K.P., et al. 1999. <strong>The</strong> DNA sequence <strong>of</strong> human chromosome22 (erratum in Nature [2000] 404: 904). Nature 402:489.Ewing B. and Green P. 1998. Base-calling <strong>of</strong> automated sequencertraces using phred. II. Error probabilities. <strong>Genom</strong>eRes. 8: 186.Ewing B., Hillier L., Wendl. M.C., and Green P. 1998. Basecalling<strong>of</strong> automated sequencer traces using phred. I. Accuracyassessment. <strong>Genom</strong>e Res. 8: 175.Farr C.J., Saiki R.K., Erlich H.A., McCormick F., and MarshallC.J. 1988. Analysis <strong>of</strong> RAS gene mutations in acute myeloidleukemia by polymerase chain reaction and oligonucleotideprobes. Proc. Natl. Acad. Sci. 85: 1629.Gonzalgo M.L. and Isaacs W.B. 2003. Molecular pathways toprostate cancer. J. Urol. 170: 2444.Gordon D., Abajian C., and Green P. 1998. Consed: A graphicaltool for sequence finishing. <strong>Genom</strong>e Res. 8: 195.Guyer B., MacDorman M.F., Martin J.A., Peters K.D., andStrobino D.M. 1998. Annual summary <strong>of</strong> vital statistics -1997. Pediatrics 102: 1333.Hamvas A., Nogee L.M., Mallory G.B., Jr., Spray T.L., HuddlestonC.B., August A., Dehner L.P., deMello D.E., Moxley M.,Nelson R., Cole F.S., and Colten H.R. 1997. Lung transplantationfor treatment <strong>of</strong> infants with surfactant protein B deficiency.J. Pediatr. 130: 231.Hattori M., Fujiyama A., Taylor T.D., Watanabe H., Yada T.,Park H.S., Toyoda A., Ishii K., Totoki Y., Choi D.K., SoedaE., Ohki M., Takagi T., Sakaki Y., Taudien S., BlechschmidtK., Polley A., Menzel U., Delabar J., Kumpf K., Lehmann R.,Patterson D., Reichwald K., Rump A., and Schillhabel M., etal. 2000. <strong>The</strong> DNA sequence <strong>of</strong> human chromosome 21. <strong>The</strong>chromosome 21 mapping and sequencing consortium. Nature405: 311.Heilig R., Eckenberg R., Petit J.L., Fonknechten N., Da Silva C.,Cattolico L., Levy M., Barbe V., de Berardinis V., Ureta-VidalA., Pelletier E., Vico V., Anthouard V., Rowen L., MadanA., Qin S., Sun H., Du H., Pepin K., Artiguenave F., RobertC., Cruaud C., Bruls T., Jaillon O., and Friedlander L., et al.2003. <strong>The</strong> DNA sequence and analysis <strong>of</strong> human chromosome14. Nature 421: 601.Hillier L.W., Fulton R.S., Fulton L.A., Graves T.A., Pepin K.H.,Wagner-McPherson C., Layman D., Maas J., Jaeger S.,Walker R., Wylie K., Sekhon M., Becker M.C., O’LaughlinM.D., Schaller M.E., Fewell G.A., Delehaunty K.D., MinerT.L., Nash W.E., Cordes M., Du H., Sun H., Edwards J.,Bradshaw-Cordum H., and Ali J., et al. 2003. <strong>The</strong> DNA sequence<strong>of</strong> human chromosome 7. Nature 424: 157.Iwai T., S. Yokota S., Nakao M., Okamoto T., Taniwaki M., OnoderaN., Watanabe A., Kikuta A., Tanaka A., Asami K.,Sekine I., Mugishima H., Nishimura Y., Koizumi S.,Horikoshi Y., Mimaya J., Ohta S., Nishikawa K., Iwai A.,Shimokawa T., Nakayama M., Kawakami K., Gushiken T.,Hyakuna N., and Fujimoto T., et al. 1999. Internal tandem duplication<strong>of</strong> the FLT3 gene and clinical evaluation in childhoodacute myeloid leukemia. <strong>The</strong> Children’s Cancer andLeukemia Study Group, Japan. Leukemia 13: 38.Kelly L.M., Yu J.C., Boulton C.L., Apatira M., Li J., SullivanC.M., Williams I., Amaral S.M., Curley D.P., Duclos N.,Neuberg D., Scarborough R.M., Pandey A., Hollenbach S.,Abeb K., Lokker N.A., Gilliland D.G., and Giese N.A. 2002.CT53518, a novel selective FLT3 antagonist for the treatment<strong>of</strong> acute myelogenous leukemia (AML). Cancer Cell 1: 421.Kiyoi H., Towatari M., Yokota S., Hamaguchi M., Ohno R.,Saito H., and Naoe T. 1998. Internal tandem duplication <strong>of</strong> theFLT3 gene is a novel modality <strong>of</strong> elongation mutation whichcauses constitutive activation <strong>of</strong> the product. Leukemia 12:1333.Kiyoi H., Naoe T., Yokota S., Nakao M., Minami S., KuriyamaK., Takeshita A., Saito K., Hasegawa S., Shimodaira S.,Tamura J., Shimazaki C., Matsue K., Kobayashi H., ArimaN., Suzuki R., Morishita H., Saito H., Ueda R., and Ohno R.1997. Internal tandem duplication <strong>of</strong> FLT3 associated withleukocytosis in acute promyelocytic leukemia. LeukemiaStudy Group <strong>of</strong> the Ministry <strong>of</strong> Health and Welfare (Kohseisho).Leukemia 11: 1447.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C.,Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W.,Funke R., Gage D., Harris K., Heaford A., Howland J., KannL., Lehoczky J., LeVine R., McEwan P., McKernan K.,Meldrim J., Mesirov J.P., Miranda C., Morris W., and NaylorJ., et al. (International Human <strong>Genom</strong>e Sequencing Consortium).2001. Initial sequencing and analysis <strong>of</strong> the humangenome. Nature 409: 860.Ley T.J., Minx P.J., Walter M.J., Ries R.E., Sun H., McLellanM., DiPersio J.F., Link D.C., Tomasson M.H., Graubert T.A.,McLeod H., Khoury H., Watson M., Shannon W., TrinkausK., Heath S., Vardiman J.W., Caligiuri M.A., BloomfieldC.D., Milbrandt J.D., Mardis E.R., and Wilson R.K. 2003. Apilot study <strong>of</strong> high-throughput, sequence-based mutationalpr<strong>of</strong>iling <strong>of</strong> primary human acute myeloid leukemia cellgenomes. Proc. Natl. Acad. Sci. 100: 14275.Manning G., Whyte D.B., Martinez R., Hunter T., and SudarsanamS. 2002. <strong>The</strong> protein kinase complement <strong>of</strong> the humangenome. Science 298: 1912.Meshinchi S., Woods W.G., Stirewalt D.L., Sweetser D.A.,Buckley J.D., Tjoa T.K., Bernstein I.D., and Radich J.P. 2001.Prevalence and prognostic significance <strong>of</strong> Flt3 internal tandemduplication in pediatric acute myeloid leukemia. Blood97: 89.Mungall A.J., Palmer S.A., Sims S.K., Edwards C.A., AshurstK.L., Wilming L., Jones M.C., Horton R., Hunt S.E., ScottC.E., Gilbert J.G., Clamp M.E., Bethel G., Milne S., AinscoughR., Almeida J.P., Ambrose K.D., Andrews T.D., AshwellR.I., Babbage A.K., Bagguley C.L., Bailey J., BanerjeeR., Barker D.J., and Barlow K.F., et al. 2003. <strong>The</strong> DNA sequenceand analysis <strong>of</strong> human chromosome 6. Nature 425:805.Nakao M., Yokota S., Iwai T., Kaneko H., Horiike S., KashimaK., Sonoda Y., Fujimoto T., and Misawa S. 1996. Internal tandemduplication <strong>of</strong> the flt3 gene found in acute myeloidleukemia. Leukemia 10: 1911.Neubauer A., Dodge R.K., George S.L., Davey F.R., Silver R.T.,Schiffer C.A., Mayer R.J., Ball E.D., Wurster-Hill D., andBloomfield C.D., et al. 1994. Prognostic importance <strong>of</strong> mutationsin the ras proto-oncogenes in de novo acute myeloidleukemia. Blood 83: 1603.Nickerson D.A., Tobe V.O., and Taylor S.L. 1997. PolyPhred:Automating the detection and genotyping <strong>of</strong> single nucleotidesubstitutions using fluorescence-based resequencing. NucleicAcids Res. 25: 2745.Nickerson D.A., Taylor S.L., Weiss K.M., Clark A.G., HutchinsonR.G., Stengard J., Salomaa V., Vartiainen E., BoerwinkleE., and Sing C.F. 1998. DNA sequence diversity in a 9.7-kbregion <strong>of</strong> the human lipoprotein lipase gene (see comments).Nat. Genet. 19: 233.Nogee L.M. 1997. Surfactant protein-B deficiency. Chest(suppl. 6) 111: 129S.Nogee L.M., de Mello D.E., Dehner L.P., and Colten H.R. 1993.Brief report: Deficiency <strong>of</strong> pulmonary surfactant protein B incongenital alveolar proteinosis. N. Engl. J. Med. 328: 406.Nogee L.M., Wert S.E., Pr<strong>of</strong>fit S.A., Hull W.M., and Whitsett J.A.2000. Allelic heterogeneity in hereditary surfactant protein B(SP-B) deficiency. Am. J. Respir. Crit. Care Med. 161: 973.Nogee L.M., Garnier G., Dietz H.C., Singer L., Murphy A.M.,deMello D.E., and Colten H.R. 1994. A mutation in the surfactantprotein B gene responsible for fatal neonatal respiratorydisease in multiple kindreds. J. Clin. Invest. 93: 1860.Radich J.P., Kopecky K.J., Willman C.L., Weick J., Head D.,Appelaum F., and Collins S.J. 1990. N-ras mutations in adultde novo acute myelogenous leukemia: Prevalence and clinicalsignificance. Blood 76: 801.
- Page 6 and 7: ForewordIn 2001, as we considered t
- Page 8: The Finished Genome Sequence of Hom
- Page 11 and 12: 4 ROGERSFigure 2. Accumulation of h
- Page 13 and 14: 6 ROGERSFigure 4. Sequencing center
- Page 15 and 16: 8 ROGERSabcFigure 5. Ensembl view o
- Page 17 and 18: 10 ROGERS2000. Analysis of vertebra
- Page 20 and 21: The Human Genome: Genes, Pseudogene
- Page 22 and 23: VARIATION ON CHROMOSOME 7 15rived f
- Page 24 and 25: VARIATION ON CHROMOSOME 7 17DNAs an
- Page 26 and 27: VARIATION ON CHROMOSOME 7 19expecte
- Page 28 and 29: VARIATION ON CHROMOSOME 7 21Drosoph
- Page 30 and 31: Mutational Profiling in the Human G
- Page 32 and 33: HUMAN MUTATIONAL PROFILING 25Anothe
- Page 36: HUMAN MUTATIONAL PROFILING 29Rieder
- Page 39 and 40: 32 SCHMUTZ ET AL.algorithm itself,
- Page 41 and 42: 34 SCHMUTZ ET AL.Figure 2. Genomic
- Page 43 and 44: 36 SCHMUTZ ET AL.compared. Some of
- Page 46 and 47: Human Subtelomeric DNAH. RIETHMAN,
- Page 48 and 49: HUMAN SUBTELOMERIC SEQUENCES 41The
- Page 50 and 51: HUMAN SUBTELOMERIC SEQUENCES 43cate
- Page 52 and 53: HUMAN SUBTELOMERIC SEQUENCES 45Figu
- Page 54: HUMAN SUBTELOMERIC SEQUENCES 47pres
- Page 57 and 58: 50 COLLINSand expand the genomics r
- Page 59 and 60: 52 COLLINSFigure 2. A public-sector
- Page 61 and 62: 54 COLLINSdefine all the parts of t
- Page 63 and 64: 56 BENTLEYmon over many generations
- Page 65 and 66: 58 BENTLEYTable 1. Genetic Disease
- Page 67 and 68: 60 BENTLEY(Clark et al. 1998; Reich
- Page 69 and 70: 62 BENTLEYACKNOWLEDGMENTSThe author
- Page 72 and 73: SNP Genotyping and Molecular Haplot
- Page 74: GENETIC ANALYSIS OF DNA POOLS 67gen
- Page 77 and 78: 70 FAN ET AL.matrix is then mated t
- Page 79 and 80: 72 FAN ET AL.Figure 3. Views of gen
- Page 81 and 82: 74 FAN ET AL.including 32 duplicate
- Page 83 and 84: 76 FAN ET AL.Figure 7. Allele-speci
- Page 85 and 86:
78 FAN ET AL.microsphere-based assa
- Page 87 and 88:
80 BERTRANPETIT ET AL.function, may
- Page 89 and 90:
82 BERTRANPETIT ET AL.diversity in
- Page 91 and 92:
84 BERTRANPETIT ET AL.gree of block
- Page 93 and 94:
86 BERTRANPETIT ET AL.Figure 1. Dec
- Page 95 and 96:
88 BERTRANPETIT ET AL.1999. Populat
- Page 97 and 98:
90 WINDEMUTH ET AL.Expression data.
- Page 99 and 100:
92 WINDEMUTH ET AL.Table 1. A Summa
- Page 101 and 102:
94 WINDEMUTH ET AL.Table 2. Signifi
- Page 103 and 104:
96 WINDEMUTH ET AL.Table 3. Summary
- Page 105 and 106:
98 WINDEMUTH ET AL.Table 6. List of
- Page 107 and 108:
100 WINDEMUTH ET AL.Table 6. (Conti
- Page 109 and 110:
102 WINDEMUTH ET AL.Table 6. (Conti
- Page 111 and 112:
104 WINDEMUTH ET AL.much of a surpr
- Page 113 and 114:
106 WINDEMUTH ET AL.Given our resul
- Page 116 and 117:
Genetic Variation and the Control o
- Page 118 and 119:
GENETIC CONTROL OF TRANSCRIPTION 11
- Page 120 and 121:
GENETIC CONTROL OF TRANSCRIPTION 11
- Page 122 and 123:
Genome-wide Detection and Analysis
- Page 124 and 125:
RECENT SEGMENTAL DUPLICATIONS 117Fi
- Page 126 and 127:
RECENT SEGMENTAL DUPLICATIONS 119St
- Page 128 and 129:
RECENT SEGMENTAL DUPLICATIONS 121Co
- Page 130 and 131:
RECENT SEGMENTAL DUPLICATIONS 123Ho
- Page 132 and 133:
The Effects of Evolutionary Distanc
- Page 134 and 135:
EVOLUTIONARY DISTANCE AND GENE PRED
- Page 136 and 137:
EVOLUTIONARY DISTANCE AND GENE PRED
- Page 138 and 139:
Lineage-specific Expansion of KRAB
- Page 140 and 141:
EVOLUTION OF ZNF GENES 133Figure 2.
- Page 142 and 143:
EVOLUTION OF ZNF GENES 135Figure 4.
- Page 144 and 145:
EVOLUTION OF ZNF GENES 137get gene,
- Page 146 and 147:
EVOLUTION OF ZNF GENES 139Y., Goodw
- Page 148 and 149:
Sequence Organization and Functiona
- Page 150 and 151:
CENTROMERE ANNOTATION 143THE CENTRO
- Page 152 and 153:
CENTROMERE ANNOTATION 145Figure 4.
- Page 154 and 155:
CENTROMERE ANNOTATION 147CONCLUSION
- Page 156:
CENTROMERE ANNOTATION 149Schueler M
- Page 159 and 160:
152 PARKHILL AND THOMSONFigure 1. T
- Page 161 and 162:
154 PARKHILL AND THOMSONshow very h
- Page 163 and 164:
156 PARKHILL AND THOMSONGene Loss a
- Page 165 and 166:
158 PARKHILL AND THOMSONYersinia ad
- Page 167 and 168:
160 MCKAY ET AL.Choosing Candidate
- Page 169 and 170:
162 MCKAY ET AL.new comparative too
- Page 171 and 172:
164 MCKAY ET AL.rich. Based on a th
- Page 173 and 174:
166 MCKAY ET AL.Embryonic Muscle an
- Page 175 and 176:
168 MCKAY ET AL.native polyadenylat
- Page 178 and 179:
Building Comparative Maps Using 1.5
- Page 180 and 181:
HUMAN CHROMOSOME 1p IN THE DOG 1731
- Page 182 and 183:
HUMAN CHROMOSOME 1p IN THE DOG 175(
- Page 184:
HUMAN CHROMOSOME 1p IN THE DOG 177l
- Page 187 and 188:
180 GEORGES AND ANDERSSON5. There i
- Page 189 and 190:
182 GEORGES AND ANDERSSONplied to r
- Page 191 and 192:
184 GEORGES AND ANDERSSONbe common
- Page 193 and 194:
186 GEORGES AND ANDERSSONin humans
- Page 196 and 197:
Evolving Methods for the Assembly o
- Page 198 and 199:
ASSEMBLING LARGE GENOMES 191Figure
- Page 200 and 201:
ASSEMBLING LARGE GENOMES 193tant ad
- Page 202 and 203:
Mouse Genome Encyclopedia ProjectY.
- Page 204 and 205:
MOUSE GENOME ENCYCLOPEDIA PROJECT 1
- Page 206 and 207:
MOUSE GENOME ENCYCLOPEDIA PROJECT 1
- Page 208 and 209:
MOUSE GENOME ENCYCLOPEDIA PROJECT 2
- Page 210 and 211:
MOUSE GENOME ENCYCLOPEDIA PROJECT 2
- Page 212 and 213:
DNA Sequence Assembly and Multiple
- Page 214 and 215:
EULERIAN ASSEMBLY AND MULTIPLE ALIG
- Page 216 and 217:
EULERIAN ASSEMBLY AND MULTIPLE ALIG
- Page 218 and 219:
EULERIAN ASSEMBLY AND MULTIPLE ALIG
- Page 220 and 221:
Ensembl: A Genome InfrastructureE.
- Page 222:
ENSEMBL 215projects often submit th
- Page 225 and 226:
218 ZHANGthe majority of these are
- Page 227 and 228:
220 ZHANGFigure 2. Demonstration of
- Page 229 and 230:
222 ZHANG(G.X. Chen et al., in prep
- Page 231 and 232:
224 ZHANGWe are waiting for experim
- Page 234 and 235:
Ontologies for Biologists: A Commun
- Page 236 and 237:
ONTOLOGIES FOR BIOLOGISTS 229al. 20
- Page 238 and 239:
ONTOLOGIES FOR BIOLOGISTS 231TOPIC
- Page 240 and 241:
ONTOLOGIES FOR BIOLOGISTS 233a.b.Fi
- Page 242:
ONTOLOGIES FOR BIOLOGISTS 2352003.
- Page 245 and 246:
238 JOSHI-TOPE ET AL.Figure 1. The
- Page 247 and 248:
240 JOSHI-TOPE ET AL.state of knowl
- Page 249 and 250:
242 JOSHI-TOPE ET AL.and co-immunop
- Page 252 and 253:
The Share of Human Genomic DNA unde
- Page 254 and 255:
DNA UNDER SELECTION FROM HUMAN-MOUS
- Page 256 and 257:
DNA UNDER SELECTION FROM HUMAN-MOUS
- Page 258 and 259:
DNA UNDER SELECTION FROM HUMAN-MOUS
- Page 260 and 261:
DNA UNDER SELECTION FROM HUMAN-MOUS
- Page 262 and 263:
Detecting Highly Conserved Regions
- Page 264 and 265:
DETECTING MULTISPECIES CONSERVED SE
- Page 266 and 267:
DETECTING MULTISPECIES CONSERVED SE
- Page 268 and 269:
DETECTING MULTISPECIES CONSERVED SE
- Page 270:
DETECTING MULTISPECIES CONSERVED SE
- Page 273 and 274:
266 ROE ET AL.noncoding regions. On
- Page 275 and 276:
268 ROE ET AL.a48 hpf embryos in Mi
- Page 277 and 278:
270 ROE ET AL.aNovel gene KIAA0819[
- Page 279 and 280:
272 ROE ET AL.aMouseRatAP00354.2 Hu
- Page 281 and 282:
274 ROE ET AL.Tautz D. and Pfeifle
- Page 283 and 284:
276 JAILLON ET AL.Detection of Evol
- Page 285 and 286:
278 JAILLON ET AL.Table 1. Distribu
- Page 287 and 288:
280 JAILLON ET AL.Table 3. Distribu
- Page 289 and 290:
282 JAILLON ET AL.ecotig is a resul
- Page 291 and 292:
284 OVCHARENKO AND LOOTSdivergent r
- Page 293 and 294:
286 OVCHARENKO AND LOOTSmodulation
- Page 295 and 296:
288 OVCHARENKO AND LOOTSsequencing
- Page 297 and 298:
290 OVCHARENKO AND LOOTSments of cl
- Page 300 and 301:
Evolution of Eukaryotic Gene Repert
- Page 302 and 303:
EVOLUTION OF EUKARYOTIC GENES AND I
- Page 304 and 305:
EVOLUTION OF EUKARYOTIC GENES AND I
- Page 306 and 307:
EVOLUTION OF EUKARYOTIC GENES AND I
- Page 308:
EVOLUTION OF EUKARYOTIC GENES AND I
- Page 311 and 312:
304 PENNACCHIO, BAROUKH, AND RUBINA
- Page 313 and 314:
306 PENNACCHIO, BAROUKH, AND RUBINh
- Page 315 and 316:
308 PENNACCHIO, BAROUKH, AND RUBINA
- Page 318:
High-Throughput Mouse Knockouts Pro
- Page 321 and 322:
314 FRIDDLE ET AL.screen to lines o
- Page 324 and 325:
Identification of Novel Functional
- Page 326 and 327:
100 bp ladder68G1168G1168H1168H6100
- Page 328 and 329:
FUNCTIONAL ELEMENTS IN HUMAN DNA 32
- Page 330 and 331:
High-resolution Human Genome Scanni
- Page 332 and 333:
HUMAN GENOME SCANNING 325false-posi
- Page 334 and 335:
HUMAN GENOME SCANNING 327affecting
- Page 336:
HUMAN GENOME SCANNING 329methods fo
- Page 339 and 340:
332 MALEK ET AL.Figure 1. The bacte
- Page 341 and 342:
334 MALEK ET AL.J., Vincent S., and
- Page 343 and 344:
336 HARDISON ET AL.reflect blocks o
- Page 345 and 346:
338 HARDISON ET AL.plain the region
- Page 347 and 348:
340 HARDISON ET AL.CALIBRATION OF T
- Page 349 and 350:
342 HARDISON ET AL.PositionRP2.3noE
- Page 351 and 352:
344 HARDISON ET AL.cific chromosoma
- Page 353 and 354:
346 WESTON ET AL.these differences
- Page 355 and 356:
348 WESTON ET AL.els controlled by
- Page 357 and 358:
350 WESTON ET AL.ures prominently i
- Page 359 and 360:
352 WESTON ET AL.nal and Bop, which
- Page 361 and 362:
354 WESTON ET AL.ablp 1466 bopbcrtB
- Page 363 and 364:
356 WESTON ET AL.like fold (Fig. 6)
- Page 366 and 367:
Implications of Genomics for Public
- Page 368 and 369:
GENETIC EPIDEMIOLOGY 361lytic epide
- Page 370 and 371:
GENETIC EPIDEMIOLOGY 363curate risk
- Page 372 and 373:
A Model System for Identifying Gene
- Page 374 and 375:
PTC TASTE GENETICS 367Figure 2. Hap
- Page 376 and 377:
PTC TASTE GENETICS 369Table 2. Hapl
- Page 378:
PTC TASTE GENETICS 371the emergence
- Page 381 and 382:
374 MCCALLION ET AL.Figure 1. Schem
- Page 383 and 384:
376 MCCALLION ET AL.lier (Carrasqui
- Page 385 and 386:
378 MCCALLION ET AL.Table 3. HSCR A
- Page 387 and 388:
380 MCCALLION ET AL.Figure 3. Trans
- Page 390 and 391:
Genetics of Schizophrenia and Bipol
- Page 392 and 393:
SCHIZOPHRENIA AND BIPOLAR AFFECTIVE
- Page 394 and 395:
SCHIZOPHRENIA AND BIPOLAR AFFECTIVE
- Page 396 and 397:
SCHIZOPHRENIA AND BIPOLAR AFFECTIVE
- Page 398 and 399:
SCHIZOPHRENIA AND BIPOLAR AFFECTIVE
- Page 400 and 401:
SCHIZOPHRENIA AND BIPOLAR AFFECTIVE
- Page 402 and 403:
The Genetics of Common Diseases: 10
- Page 404 and 405:
GENETICS OF COMMON DISEASES 397with
- Page 406 and 407:
GENETICS OF COMMON DISEASES 399SELE
- Page 408:
GENETICS OF COMMON DISEASES 401F.,
- Page 411 and 412:
404 CHEUNG ET AL.netic analysis. Ex
- Page 413 and 414:
406 CHEUNG ET AL.Figure 3. The expr
- Page 416 and 417:
Regulation of α-Synuclein Expressi
- Page 418 and 419:
α-SYNUCLEIN EXPRESSION AND PD 411T
- Page 420 and 421:
1. The levels of α-synuclein prote
- Page 422:
α-SYNUCLEIN EXPRESSION AND PD 415g
- Page 425 and 426:
418 BOTSTEINFigure 1. (A) Blectron
- Page 427 and 428:
420 BOTSTEINFigure 3. Cluster diagr
- Page 429 and 430:
422 BOTSTEINFigure 6. Kaplan-Meier
- Page 431 and 432:
424 BOTSTEINGarber M.E., Troyanskay
- Page 433 and 434:
426 ANTONARAKIS ET AL.1316192225283
- Page 435 and 436:
428 ANTONARAKIS ET AL.Figure 5. Sam
- Page 437 and 438:
430 ANTONARAKIS ET AL.POPULATION VA
- Page 439 and 440:
432 JORGENSEN ET AL.tive small mole
- Page 441 and 442:
434 JORGENSEN ET AL.FLAG-tagged pro
- Page 443 and 444:
436 JORGENSEN ET AL.visualization t
- Page 445 and 446:
438 JORGENSEN ET AL.AArp2/3 Complex
- Page 447 and 448:
Pathway40S440 JORGENSEN ET AL.ANutr
- Page 449 and 450:
442 JORGENSEN ET AL.Giaever G., Chu
- Page 452 and 453:
Genomic Disorders: Genome Architect
- Page 454 and 455:
GENOME ARCHITECTURE AND GENOMIC DIS
- Page 456 and 457:
GENOME ARCHITECTURE AND GENOMIC DIS
- Page 458 and 459:
GENOME ARCHITECTURE AND GENOMIC DIS
- Page 460 and 461:
GENOME ARCHITECTURE AND GENOMIC DIS
- Page 462 and 463:
Human Versus Chimpanzee Chromosome-
- Page 464 and 465:
HUMAN VS. CHIMP CHROMOSOME COMPARIS
- Page 466 and 467:
HUMAN VS. CHIMP CHROMOSOME COMPARIS
- Page 468 and 469:
Novel Transcriptional Units and Unc
- Page 470 and 471:
TRANSCRIPTIONAL UNITS AND GENE PAIR
- Page 472 and 473:
TRANSCRIPTIONAL UNITS AND GENE PAIR
- Page 474 and 475:
TRANSCRIPTIONAL UNITS AND GENE PAIR
- Page 476 and 477:
TRANSCRIPTIONAL UNITS AND GENE PAIR
- Page 478 and 479:
mtDNA Variation, Climatic Adaptatio
- Page 480 and 481:
mtDNA VARIATION 473Figure 3. Region
- Page 482 and 483:
ANALYSIS OF ADAPTIVE SELECTION FORR
- Page 484 and 485:
mtDNA VARIATION 477Figure 8. Temper
- Page 486 and 487:
Positive Selection in the Human Gen
- Page 488 and 489:
HUMAN-SPECIFIC EVOLUTIONARY CHANGES
- Page 490 and 491:
HUMAN-SPECIFIC EVOLUTIONARY CHANGES
- Page 492:
HUMAN-SPECIFIC EVOLUTIONARY CHANGES
- Page 495 and 496:
488 UNDERHILLorigin episodes, each
- Page 497 and 498:
490 UNDERHILLhaplogroups C through
- Page 499 and 500:
492 UNDERHILLO (Fig. 2e) that share
- Page 502 and 503:
The New Quantitative BiologyM.V. OL
- Page 504 and 505:
NEW QUANTITATIVE BIOLOGY 497alone.
- Page 506 and 507:
NEW QUANTITATIVE BIOLOGY 499There w
- Page 508 and 509:
NEW QUANTITATIVE BIOLOGY 501ceded,