conservation across taxa: divided strainsand united kingdoms. Nucleic Acids Res2005a;33(2):616-21.Kunin V, Goldovsky L, Darzentas N,Ouzounis CA. The net of life:reconstructing the microbialphylogenetic network. Genome Res2005b Jul;15(7):954-9.Kurtz S, Phillippy A, Delcher AL, SmootM, Shumway M, Antonescu C, et al.Versatile and open software forcomparing large genomes. Genome Biol2004;5(2):R12.Lee D, Redfern O, Orengo C. Predictingprotein function from sequence andstructure. Nat Rev Mol Cell Biol 2007December;8(12):995-1005.Legault B, Lopez-Lopez A, ba-CasadoJ, Doolittle WF, Bolhuis H, Rodriguez-Valera F, Papke RT. Environmentalgenomics of “Haloquadratum walsbyi”in a saltern crystallizer indicates a largepool of accessory genes in an otherwisecoherent species. BMC Genomics2006;7(1):171.Lerat E, Ochman H. Recognizing thepseudogenes in bacterial genomes.Nucleic Acids Res 2005;33(10):3125-3132.Lipman DJ, Pearson WR. Rapid andsensitive protein similarity searches.Science 1985 Mar 22;227(4693):1435-41.Liu Y, Harrison PM, Kunin V, GersteinM. Comprehensive analysis ofpseudogenes in prokaryotes:widespread gene decay and failure ofputative horizontally transferred genes.Genome Biol 2004;5(9):R64.Ma B, Tromp J, Li M. PatternHunter:faster and more sensitive homologysearch. Bioinformatics 2002Mar;18(3):440-5.Medini D, Donati C, Tettelin H,Masignani V, Rappuoli R. The microbialpan-genome. Current Opinion inGenetics & Development 2005December;15(6):589-594.Moran NA. Microbial minimalism:genome reduction in bacterialpathogens. Cell 2002 March 8;108(5):583-586.Morett E, Korbel JO, Rajan E, Saab-Rincon G, Olvera L, Olvera M, SchmidtS, Snel B, Bork P. Systematic discoveryof analogous enzymes in thiaminbiosynthesis. Nat Biotechnol. 2003,21(7):790-5.Morgenstern B, Frech K, Dress A, WernerT. DIALIGN: finding local similarities bymultiple sequence alignment.Bioinformatics 1998;14(3):290-4.Morgenstern B, Rinner O, AbdeddaimS, Haase D, Mayer KF, Dress AW, et al.Exon discovery by genomic sequencealignment. Bioinformatics 2002Jun;18(6):777-87.Morgenstern B. DIALIGN 2:improvement of the segment-to-segmentapproach to multiple sequencealignment. Bioinformatics 1999Mar;15(3):211-8.Moya A, Pereto J, Gil R, Latorre A.Learning how to live together: genomicinsights into prokaryote-animalsymbioses. Nat Rev Genet 2008March;9(3):218-229.Needleman SB, Wunsch CD. A generalmethod applicable to the search forsimilarities in the amino acid sequenceof two proteins. J Mol Biol 1970Mar;48(3):443-53.Ochman H, Davalos LM. The nature anddynamics of bacterial genomes. Science2006 March 24;311(5768):1730-1733.Ochman H, Moran NA. Genes lost andgenes found: evolution of bacterialpathogenesis and symbiosis. Science2001 May 11;292(5519):1096-1099.Otu HH, Sayood K. A new sequencedistance measure for phylogenetic treeconstruction. Bioinformatics 2003 Nov1;19(16):2122-30.Pareja E, Pareja-Tobes P, Manrique M,Pareja-Tobes E, Bonal J, Tobes R.ExtraTrain: a database of Extragenicregions and Transcriptional informationin prokaryotic organisms. BMCMicrobiol 2006;6:29.Passos GAS, Nguyen C, Jordan B. ProjetoTranscriptoma: Análise da ExpressãoGênica em Larga Escala UsandoDNA - Arrays. Biotecnol Ciênc Des 2000Janeiro/Fevereiro;12:34-<strong>37</strong>.Patterson SD, Aebersold RH. Proteomics:the first decade and beyond. Nat Genet2003 March;33 Suppl:311-323.Pearson WR, Lipman DJ. Improved toolsfor biological sequence comparison.Proc Natl Acad Sci U S A 1988Apr;85(8):2444-8.Peregrin-Alvarez JM, Tsoka S, OuzounisCA. The phylogenetic extent ofmetabolic enzymes and pathways.Genome Res 2003 March;13(3):422-427.Prosdocimi F, Cerqueira GC, Binneck E,Silva AF, Reis AN, Junqueira ACM, et al.Bioinformática: Manual do Usuário - Umguia básico e amplo sobre os diversosaspectos dessa nova. Biotecnol CiêncDes 2002 Novembro/Dezembro;29:12-25.Randhawa GS, Bishai WR. Beneficialimpact of genome projects ontuberculosis control. Infect Dis ClinNorth Am 2002 Mar;16(1):145-61.Rigden DJ, Mello LV. Anotação funcionalcomputacional de proteínas: Novosmétodos computacionais poderão preencherlacunas do sistema de anotaçãoatual. Biotecnol Ciênc Des 2002 Março/Abril;25:64-70.Rocha EP. Order and disorder in bacterialgenomes. Curr Opin Microbiol 2004bOctober;7(5):519-527.Rocha EP. The replication-relatedorganization of bacterial genomes.Microbiology 2004a June;150(Pt 6):1609-1627.Schwartz S, Kent WJ, Smit A, Zhang Z,Baertsch R, Hardison RC, et al. Humanmousealignments with BLASTZ. GenomeRes 2003 Jan;13(1):103-7.Shendure JA, Porreca GJ, Church GM.Overview of DNA sequencing strategies.Curr Protoc Mol Biol 2008 January;Chapter7:Unit.Skrabanek L, Saini HK, Bader GD, EnrightAJ. Computational prediction of proteinproteininteractions. Mol Biotechnol 2008January;38(1):1-17Smith TF, Waterman MS. Comparison ofBiosequences. Adv. Appl. Math. 1981; 2:482-9.Souza MV, Fontes W, Ricart CAO. Análisede Proteomas: O despertar da era pósgenômica.Biotecnol Ciênc Des 1999 Janeiro/Fevereiro;7:12-14.Stein L. Genome annotation: fromsequence to biology. Nat Rev Genet 2001July;2(7):493-503.Strohman RC. The coming Kuhnianrevolution in biology. Nat Biotechnol 1997Mar;15(3):194-200.Tekaia F, Yeramian E. Genome trees fromconservation profiles. PLoS Comput Biol2005 Dec;1(7):e75.Tettelin H, Masignani V, Cieslewicz MJ,Donati C, Medini D, Ward NL, Angiuoli SV,Crabtree J, Jones AL, Durkin AS, DeBoyRT, Davidsen TM, Mora M, Scarselli M, RosI, Peterson JD, Hauser CR, Sundaram JP,Nelson WC, Madupu R, Brinkac LM,Dodson RJ, Rosovitz MJ, Sullivan SA,Daugherty SC, Haft DH, Selengut J, GwinnML, Zhou L, Zafar N, Khouri H, Radune D,Dimitrov G, Watkins K, O’Connor KJB,Smith S, Utterback TR, White O, RubensCE, Grandi G, Madoff LC, Kasper DL,Telford JL, Wessels MR, Rappuoli R, FraserCM. Genome analysis of multiplepathogenic isolates of Streptococcusagalactiae: Implications for the microbial“pan-genome”. Proceedings of the NationalAcademy of Sciences 2005 September27;102(39):13950-13955.Thompson JD, Higgins DG, Gibson TJ.CLUSTAL W: improving the sensitivity ofprogressive multiple sequence alignmentthrough sequence weighting, positionspecificgap penalties and weight matrixchoice. Nucleic Acids Res 1994 Nov11;22(22):4673-80.Wei L, Liu Y, Dubchak I, Shon J, Park J.Comparative genomics approaches to studyorganism similarities and differences. JBiomed Inform 2002 Apr;35(2):142-50.28 <strong>Biotecnologia</strong> Ciência & <strong>Desenvolvimento</strong> - nº <strong>37</strong>
GLOSSÁRIOAlgoritmo. Procedimento organizado (passos e instruções)para executar um determinado tipo de cálculoou solucionar um determinado tipo de problema.Alinhamento de seqüências. Processo de alinhar (colocarlado a lado) duas ou mais seqüências do mesmotipo (nucleotídicas ou protéicas) de forma a obter omáximo de identidade entre elas com o propósito dedeterminar o grau de similaridade.Alinhamento global. Alinhamento de pares de seqüênciasnucleotídicas ou protéicas ao longo de todaa extensão das mesmas.Alinhamento local. Alinhamento de uma ou maispartes de duas seqüências nucleotídicas ou protéicas.Análise filogenética. Análise filogenética ou filogeniaconsiste no estudo das relações evolutivas (ouseja, na reconstrução da história evolutiva) entre gruposde organismos ou outras entidades que se acreditapossuírem um ancestral comum, como por exemplo,espécies, populações e genes.Analogia. Relação entre dois caracteres quaisquer quedescendem, por convergência, de caracteres ancestraisnão relacionados entre si (Fitch 1970, 2000).Bioinformática e Biologia Computacional. Em 17 dejulho de 2000, o National Institutes of Health (NIH),uma das agências do departamento de saúde norteamericanocom reconhecimento internacional na áreade pesquisa biomédica, divulgou sua definição detrabalho para Bioinformática e para Biologia Computacional,elaborada pelo Biomedical Information Scienceand Technology Initiative Consortium (BISTIC)Definition Committee. De acordo com este documento“A bioinformática e a biologia computacional têmsuas raízes nas ciências da vida bem como nasciências da computação e informação e na tecnologia.Ambas estas abordagens interdisciplinaresse beneficiam de disciplinas específicas, tais comoa matemática, a física, as ciências da computaçãoe a engenharia, a biologia e as ciências docomportamento. Cada uma delas mantém interaçõesmuito estreitas com as ciências da vida paraconcretizar todo o seu potencial. A bioinformáticaaplica princípios das ciências da informação e datecnologia para tornar os vastos, diversificados ecomplexos dados produzidos pelas ciências da vidamais compreensíveis e úteis. A biologia computacionalusa abordagens matemáticas e computacionaispara resolver questões teóricas e experimentaisna biologia. Embora a bioinformática e a biologiacomputacional sejam distintas, há significativasobreposição e atividade em suas interfaces.(...) Bioinformática: pesquisa, desenvolvimento ouaplicação de ferramentas e abordagens computacionaispara ampliar o uso de dados de origembiológica, médica, comportamental ou de saúde,incluindo adquirir, armazenar, organizar, arquivar,analisar ou visualizar tais dados. BiologiaComputacional: desenvolvimento e aplicação demétodos analíticos e teóricos de dados e técnicasde modelagem matemática e simulação computacionalpara o estudo de sistemas biológicos, comportamentaise sociais.” (BISTIC Definition Committee,2000). [Tradução livre do autor].DNA. Sigla em inglês para deoxyribonucleic acid, ouácido desoxirribonucléico. Ácido nucléico constituídopor desoxirribose, fosfato e pelas bases nitrogenadasadenina, guanina, citosina e timina. Contém asinstruções genéticas usadas no desenvolvimento e funcionamentode todos os seres vivos.Fatores epigenéticos. Fatores (não genéticos) responsáveispelo controle temporal e espacial da atividadede todos os genes necessários para o desenvolvimentode um organismo complexo desde o zigoto até a faseadulta (citado por Strohman 1997).Fusão/Fissão gênica. Foi observado que determinadospares de proteínas funcionalmente relacionadasentre si, presentes em certos organismos, têm homólogosem outros organismos fundidos em uma únicacadeia protéica (Marcotte et al 1999; Enright et al 1999).O processo de formação destas proteínas é chamadode fusão gênica (quando há a adição de genes ouseqüências funcionais em uma cadeia de DNA) oufissão gênica (quando há a perda de genes ou seqüênciasfuncionais em uma cadeia de DNA). Eventos defusão/fissão gênica são fenômenos naturais reconhecidoscomo uma das principais forças evolutivasna criação de proteínas de múltiplos domínios.Genes. Genes são as unidades hereditárias emtodos os organismos vivos, formando componentesessenciais do genoma (o conjunto completode informação genética) destes organismos,sendo responsáveis pelo desenvolvimento físico,pelo metabolismo e também, até certo ponto,seu comportamento. Alguns genes produzemmoléculas de RNA enquanto outros desempenhamimportantes papéis regulatórios ou estruturais.A maioria dos genes codifica proteínas,grandes moléculas compostas de longas cadeiasde aminoácidos, que respondem pela maioriadas reações químicas desempenhadas pela célula.Genoma. Termo criado, em 1920, por Hans Winkler,professor de Botânica na Universidade deHamburgo. Designa toda a informação hereditáriade um organismo que está codificada no seuDNA (ou, em alguns vírus, no RNA). Isto incluitanto os genes como as seqüências não codificadoras(conhecidas como DNA-lixo).Genômica. Análise (em larga escala) do genomacompleto de um organismo.Homologia. Relação entre dois caracteres (traçosgenéticos, estruturais ou funcionais de umorganismo) quaisquer que descendem de um caractereancestral comum, normalmente com divergência(Fitch 1970, 2000).Matriz de substituição. Matriz que representatodas as possíveis trocas entre aminoácidos, nasquais um valor é atribuído a cada uma destastrocas. Estes valores são proporcionais à probabilidadede ocorrência de cada troca, tomandosecomo base um determinado modelo evolutivo.PAM – Percent Accepted Mutation (Dayhoffet al 1978). BLOSUM - BLOcks SUbstitution Matrix(Henikoff & Henikoff 1992).Metagenômica. Também conhecida como genômicaambiental, ecogenômica, ou ainda genômicade comunidades, consiste na análise domaterial genético obtido diretamente de amostrasambientais, permitindo o estudo de organismosque não podem ser facilmente cultivadosem laboratório, bem como o estudo de organismosem seus ambientes naturais (Metagenomics2008).Micobactérias. O gênero Mycobacterium (familiaMycobacteriaceae, ordem Actinomycetales),um dos mais antigos e bem conhecidos gênerosde bactéria, foi introduzido por Lehmann e Neumannem 1896, para incluir os agentes causadoresda hanseníase e da tuberculose, bactérias quehaviam sido anteriormente classificadas comoBacterium leprae e Bacterium tuberculosis, respectivamente(Goodfellow & Minnikin, 1984).Os organismos pertencentes a este gênero sãoaeróbios, imóveis e não formam endósporos ouesporos; têm forma de bastonetes delgados, retosou ligeiramente encurvados, com raras formasramificadas. Seu DNA é rico em guanina (G) ecitosina (C) (de 62 a 70% G+C, com exceção deMycobacterium leprae que tem 57.8% de GC). Asmicobactérias possuem ainda características peculiarescomo álcool-ácido resistência (uma vezcoradas por corantes básicos, resistem à descoloraçãopor soluções álcool-ácidas sendo, portanto,denominadas bacilos álcool-ácido resistentes)e resistência incomum à dessecação e a agentesquímicos.Ortólogos. Genes homólogos em espécies diferentesoriginados de um gene ancestral comum,durante a especiação (Fitch 1970, 2000).Óperon. Grupo de genes funcionalmente relacionadosentre si, regulados (em conjunto) por ummesmo operador.Parálogos. Genes homólogos em uma espécieem particular originados por duplicação (Fitch1970, 2000).Proteínas. Moléculas compostas por aminoácidosligados entre si em uma ordem particular,especificada pelas seqüências de DNA dos genesque as codificam. São componentes essenciais dos organismos,participando de todos os processos celulares(catálise enzimática, sinalização celular, resposta imune,adesão celular, ciclo celular etc.) e também comocomponentes estruturais e mecânicos.Proteômica. Análise (em larga escala) do conjunto completode proteínas expressas por uma célula, tecido ouorganismo, em um dado momento e sob certas circunstânciasambientais.Regiões sintênicas. Sintenia foi um termo originalmentecunhado para designar a presença de dois ou maisloci gênicos (próximos ou não) no mesmo cromossomo.Atualmente, refere-se também a duas regiões degenomas distintos que mostram considerável grau desimilaridade de seqüência entre si e algum grau de conservaçãoda ordem dos genes nestas regiões e que, portanto,têm probabilidade de descender de um ancestralcomum.Regiões de baixa complexidade. Regiões em ácidosnucléicos ou proteínas com desvios na composição deseus resíduos (nucleotídeos ou aminoácidos), incluindotratos homopoliméricos (longas seqüências formadaspelo mesmo resíduo), repetições com curtos espaçosentre si e sobre-representações mais sutis de algunsresíduos.Seqüência genômica. Toda ou parte da cadeia de DNAque compõe um genoma.Simbiontes. Organismos que vivem em simbiose, isto é,dois organismos distintos que mantêm íntima e longaassociação entre si, na qual um ou ambos se beneficiamdesta relação. Esta associação inclui relações nas quaisuma das partes vive sobre (ectobiose) ou dentro (endobiose)da outra, podendo ser obrigatória, ou seja, necessáriaà sobrevivência de pelo menos um dos organismosenvolvidos, ou facultativa, na qual a associação ébenéfica, porém não é essencial à sobrevivência dosorganismos. As categorias de simbiose incluem o mutualismo(quando ambos se beneficiam), o comensalismo(quando apenas um se beneficia e o outro não é significativamentelesado ou beneficiado) e o parasitismo(quando apenas um se beneficia e o outro é lesado pelarelação) (Symbiosis 2008).Transcriptômica. Análise (em larga escala) do conjuntode todos os RNA mensageiros (transcritos) de umacélula, tecido ou organismo, em um dado momento esob certas circunstâncias ambientais.Referências bibliográficasBISTIC Definition Committee. NIH working definition ofbioinformatics and computational biology. 2000. Disponívelem: Acesso em: 26 mar. 2008.Dayhoff MO, Schwartz RM, Orcutt BC. A model of evolutionarychange in proteins. In: Dayhoff MO, ed. Atlas ofProtein Sequence and Structure. Washington DC: NationalBiomedical Research Foundation; 1978. v.5. Suppl.3.p.345-352.Enright AJ, Iliopoulos I, Kyrpides NC, Ouzounis CA.Protein interaction maps for complete genomes basedon gene fusion events. Nature 1999 Nov 4;402(6757):86-90.Fitch WM. Distinguishing homologous from analogousproteins. Syst Zool 1970 Jun;19(2):99-113.Fitch WM. Homology a personal view on some of theproblems. Trends Genet 2000 May;16(5):227-31.Goodfellow M, Minnikin DE. Circunscription of the genus.In: Kubica GP, Wayne LG, eds. The Mycobacteria: ASource Book. New York: Marcel Dekker; 1984. p.1-24.Henikoff S, Henikoff JG. Amino acid substitution matricesfrom protein blocks. Proc Natl Acad Sci U S A 1992Nov 15;89(22):10915-9.Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO,Eisenberg D. Detecting protein function and proteinproteininteractions from genome sequences. Science1999 Jul 30;285(5428):751-3.Metagenomics. In Wikipedia: The Free Encyclopedia.Wikimedia Foundation Inc. Encyclopedia on-line. Disponívelem: Acesso em: 26 mar. 2008.Strohman RC. The coming Kuhnian revolution in biology.Nat Biotechnol 1997 Mar;15(3):194-200.Symbiosis. In Wikipedia: The Free Encyclopedia. WikimediaFoundation Inc. Encyclopedia on-line. Disponívelem: Acessoem: 26 mar. 2008.<strong>Biotecnologia</strong> Ciência & <strong>Desenvolvimento</strong> - nº <strong>37</strong> 29
- Page 1 and 2: Biotecnologia Ciência & Desenvolvi
- Page 3 and 4: BC&D - Como o senhor avalia aposiç
- Page 5 and 6: dicamentos.Os anticorpos monoclonai
- Page 8 and 9: PesquisaLUCIFERASES DE VAGALUMESPes
- Page 10 and 11: ger & McElroy, 1965; Hastings,1995)
- Page 12 and 13: ciferases de elaterídeos e fengod
- Page 14 and 15: utilizados como marcadores sensíve
- Page 16: 5,5-dimethyloxyluciferin. JACS 124:
- Page 19 and 20: Rev. Bras. Entom. 33: 359-366..Vivi
- Page 21 and 22: vimento de pesquisas científicas (
- Page 23 and 24: ubíquos) é inferior a 100, envolv
- Page 25 and 26: (tais como genes, operons, grupos[c
- Page 27: evolution of mycobacterialpathogeni
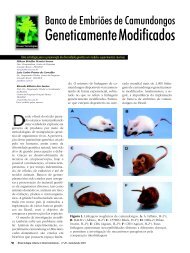
- Page 31 and 32: oócito e melhora o posterior desen
- Page 33 and 34: Figura 7: Ilustração de resultado
- Page 35 and 36: 10. COELHO, L. A.; ESPER, C. R.;GAR
- Page 37 and 38: - permitir a rápida liberação do
- Page 39 and 40: de crescimento, quatro grupos de mi
- Page 41 and 42: skovia xanthineolytica LL-G109 (And
- Page 43 and 44: Tabela 1. Purificação da β-1,3 g
- Page 45 and 46: depende em parte, da venda dos subp
- Page 47 and 48: tipo poro-canal-G1pF atua por sensi
- Page 49 and 50: eutilização de células imobiliza
- Page 51 and 52: ANP - AGENCIA NACIONAL DE PETROLEO
- Page 53 and 54: inantes, incluindo eritropoetinas,
- Page 55 and 56: proteínas terapêuticas como, por
- Page 57 and 58: capsuladas, a redução de efeitos
- Page 59 and 60: Zhang W, Rong Z, Chen H, Jiang X.Br
- Page 61 and 62: FIGURA 1. Estrutura “Markush”Ma
- Page 63 and 64: Figura 2, todos os compostos origin
- Page 65 and 66: Figura 2. O filo Porifera é dividi
- Page 67 and 68: Figura 4. Microscopia eletrônica d
- Page 69 and 70: plication of genomics to uncultured
- Page 71 and 72: viabilidade celular (1) a técnica
- Page 73 and 74: Gráfico 02: Média das viabilidade
- Page 75 and 76: Tabela 04: Análise das médias de
- Page 77 and 78: icus blazei. Atualmente é também
- Page 79:
5. Agaricus sylvaticus é sinônimo