The Origin and Evolution of Mammals - Moodle
The Origin and Evolution of Mammals - Moodle
The Origin and Evolution of Mammals - Moodle
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
276 THE ORIGIN AND EVOLUTION OF MAMMALS<br />
82<br />
81<br />
78<br />
77<br />
Cetartiodactyla<br />
76<br />
Eulipotyphla<br />
Chiroptera<br />
Rodentia<br />
55<br />
56<br />
57<br />
58<br />
50<br />
51<br />
Primates<br />
49<br />
Xenarthra<br />
65<br />
66<br />
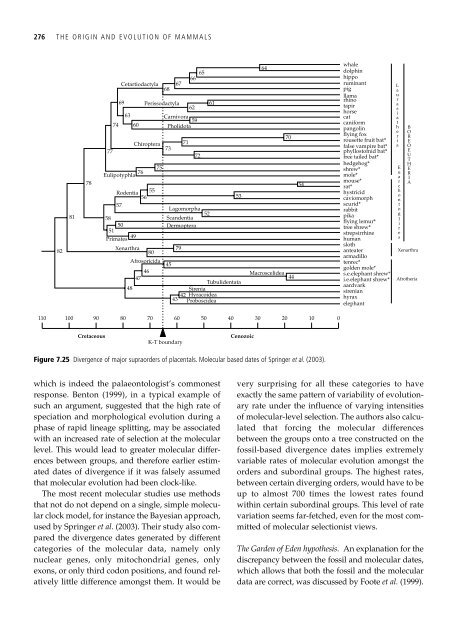
Figure 7.25 Divergence <strong>of</strong> major supraorders <strong>of</strong> placentals. Molecular based dates <strong>of</strong> Springer et al. (2003).<br />
which is indeed the palaeontologist’s commonest<br />
response. Benton (1999), in a typical example <strong>of</strong><br />
such an argument, suggested that the high rate <strong>of</strong><br />
speciation <strong>and</strong> morphological evolution during a<br />
phase <strong>of</strong> rapid lineage splitting, may be associated<br />
with an increased rate <strong>of</strong> selection at the molecular<br />
level. This would lead to greater molecular differences<br />
between groups, <strong>and</strong> therefore earlier estimated<br />
dates <strong>of</strong> divergence if it was falsely assumed<br />
that molecular evolution had been clock-like.<br />
<strong>The</strong> most recent molecular studies use methods<br />
that not do not depend on a single, simple molecular<br />
clock model, for instance the Bayesian approach,<br />
used by Springer et al. (2003). <strong>The</strong>ir study also compared<br />
the divergence dates generated by different<br />
categories <strong>of</strong> the molecular data, namely only<br />
nuclear genes, only mitochondrial genes, only<br />
exons, or only third codon positions, <strong>and</strong> found relatively<br />
little difference amongst them. It would be<br />
68<br />
67<br />
69 Perissodactyla<br />
62<br />
74<br />
63<br />
60<br />
Carnivora<br />
59<br />
Pholidota<br />
80<br />
75<br />
73<br />
71<br />
72<br />
Lagomorpha<br />
Sc<strong>and</strong>entia<br />
Dermoptera<br />
52<br />
79<br />
Afrosoricida<br />
45<br />
46<br />
47<br />
Tubulidentata<br />
48<br />
Sirenia<br />
42 Hyracoidea<br />
43 Proboscidea<br />
110 100 90 80 70 60 50 40 30 20 10 0<br />
Cretaceous<br />
K-T boundary<br />
61<br />
53<br />
Cenozoic<br />
64<br />
Macroscelidea<br />
70<br />
44<br />
54<br />
whale<br />
dolphin<br />
hippo<br />
ruminant<br />
pig<br />
llama<br />
rhino<br />
tapir<br />
horse<br />
cat<br />
caniform<br />
pangolin<br />
flying fox<br />
rousette fruit bat*<br />
false vampire bat*<br />
phyllostomid bat*<br />
free tailed bat*<br />
hedgehog*<br />
shrew*<br />
mole*<br />
mouse*<br />
rat*<br />
hystricid<br />
caviomorph<br />
scurid*<br />
rabbit<br />
pika<br />
flying lemur*<br />
tree shrew*<br />
strepsirrhine<br />
human<br />
sloth<br />
L<br />
aurasiatheria<br />
E<br />
u<br />
a<br />
r<br />
c<br />
h<br />
o<br />
n<br />
t<br />
o<br />
g<br />
l<br />
i<br />
r<br />
e<br />
s<br />
B<br />
O<br />
RE<br />
O<br />
EUT<br />
H<br />
E<br />
R<br />
I<br />
A<br />
anteater<br />
armadillo<br />
tenrec*<br />
golden mole*<br />
s.e.elephant shrew*<br />
Xenarthra<br />
i.e.elephant shrew*<br />
aardvark<br />
sirenian<br />
hyrax<br />
elephant<br />
Afrotheria<br />
very surprising for all these categories to have<br />
exactly the same pattern <strong>of</strong> variability <strong>of</strong> evolutionary<br />
rate under the influence <strong>of</strong> varying intensities<br />
<strong>of</strong> molecular-level selection. <strong>The</strong> authors also calculated<br />
that forcing the molecular differences<br />
between the groups onto a tree constructed on the<br />
fossil-based divergence dates implies extremely<br />
variable rates <strong>of</strong> molecular evolution amongst the<br />
orders <strong>and</strong> subordinal groups. <strong>The</strong> highest rates,<br />
between certain diverging orders, would have to be<br />
up to almost 700 times the lowest rates found<br />
within certain subordinal groups. This level <strong>of</strong> rate<br />
variation seems far-fetched, even for the most committed<br />
<strong>of</strong> molecular selectionist views.<br />
<strong>The</strong> Garden <strong>of</strong> Eden hypothesis. An explanation for the<br />
discrepancy between the fossil <strong>and</strong> molecular dates,<br />
which allows that both the fossil <strong>and</strong> the molecular<br />
data are correct, was discussed by Foote et al. (1999).